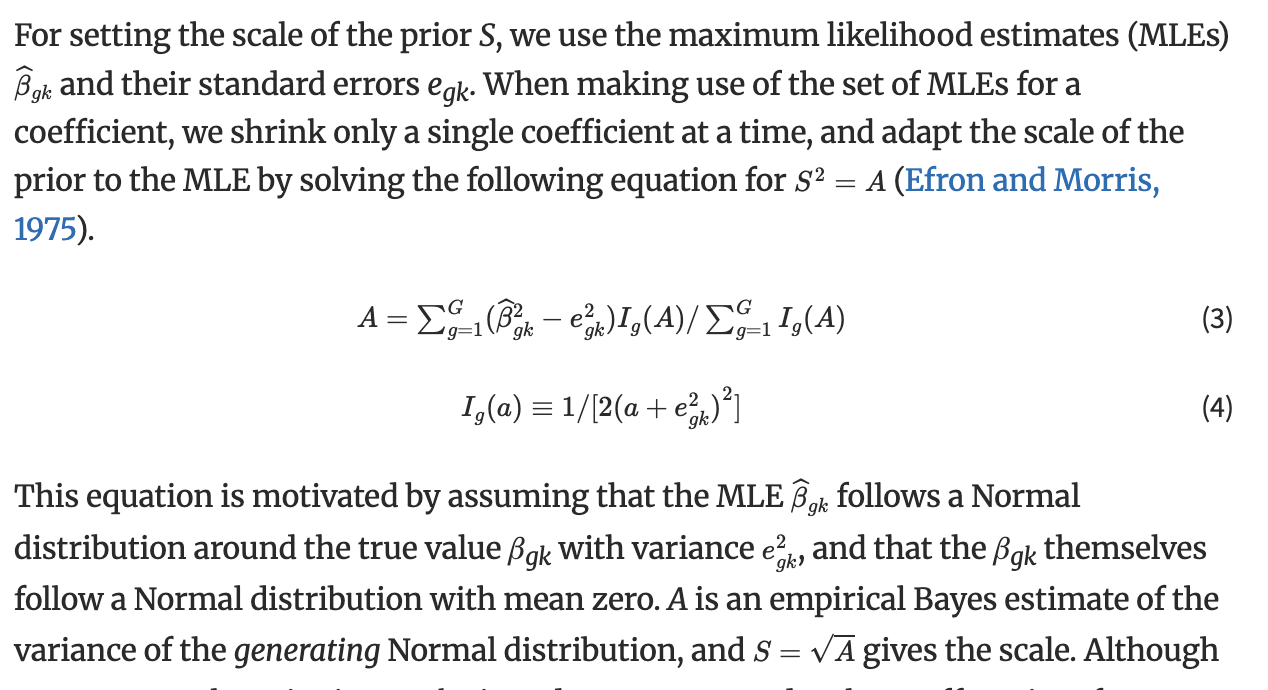
Hello,
I'm reading the DESeq2 paper, and i have two questions regarding the prior for LFC.
1) For the variance of the normal prior for LFC, I am wondering why the variance contributed by sampling distribution of LFC isn't separated out, unlike the prior for log dispersion.
2) The model's coefficients (LFC's) estimates for a gene should be correlated, while the priors for them are set as independent. Is there a specific reason for this choice? Wouldn't a multivariate normal prior be more appropriate here?
Any help is greatly appreciated.