Dear List
I obtained a non-monotonic Voom plot with voomByGroup on Xenium data. Does this compromise the reliability of the results, and if so is there anything I can do about it? Here's the plot
Here is my code up until an including the plot and sessionInfo.
library(Seurat)
library(limma)
library(edgeR)
system("mkdir results/")
system("mkdir results/global/")
source("voomByGroup.R")
targets<-readTargets("data/targets.txt")
sprsmat.tmp=Read10X_h5(paste0("data/",targets[1,1]))
counts= rowSums(sprsmat.tmp)
for (i in 2:nrow(targets)){
region.tmp.sprsmat=Read10X_h5(paste0("data/",targets[i,1]))
counts.tmp=rowSums(region.tmp.sprsmat)
counts=cbind(counts,counts.tmp)
}
colnames(counts)<-targets$Name
flevels<-unique(targets$Target)
f<-factor(targets$Target,levels=flevels)
y<-DGEList(counts=counts, genes=rownames(counts))
isexpr<-rowSums(y$counts>=10) >= 3
hasannot<-rowSums(is.na(y$genes))==0
y<-y[isexpr & hasannot,]
y$samples$lib.size<- colSums(y$counts)
y$samples$lib.size
y<-calcNormFactors(y)
des<-model.matrix(~0+f)
colnames(des)<-flevels
png("results/global/voomByGrouplot.png")
v=voomByGroup(y, design=des, group=targets$Target,plot="combine")
dev.off()
> sessionInfo()
R version 4.5.1 (2025-06-13)
Platform: x86_64-apple-darwin20
Running under: macOS Sonoma 14.6.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.5-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.5-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.1
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] edgeR_4.6.3 limma_3.64.1 Seurat_5.3.0 SeuratObject_5.1.0
[5] sp_2.2-0
loaded via a namespace (and not attached):
[1] deldir_2.0-4 pbapply_1.7-2 gridExtra_2.3
[4] rlang_1.1.6 magrittr_2.0.3 RcppAnnoy_0.0.22
[7] matrixStats_1.5.0 ggridges_0.5.6 compiler_4.5.1
[10] spatstat.geom_3.4-1 png_0.1-8 vctrs_0.6.5
[13] reshape2_1.4.4 hdf5r_1.3.12 stringr_1.5.1
[16] pkgconfig_2.0.3 fastmap_1.2.0 promises_1.3.3
[19] bit_4.6.0 purrr_1.1.0 jsonlite_2.0.0
[22] goftest_1.2-3 later_1.4.2 spatstat.utils_3.1-4
[25] irlba_2.3.5.1 parallel_4.5.1 cluster_2.1.8.1
[28] R6_2.6.1 ica_1.0-3 stringi_1.8.7
[31] RColorBrewer_1.1-3 spatstat.data_3.1-6 reticulate_1.42.0
[34] parallelly_1.45.0 spatstat.univar_3.1-4 lmtest_0.9-40
[37] scattermore_1.2 Rcpp_1.1.0 tensor_1.5.1
[40] future.apply_1.20.0 zoo_1.8-14 sctransform_0.4.2
[43] httpuv_1.6.16 Matrix_1.7-3 splines_4.5.1
[46] igraph_2.1.4 tidyselect_1.2.1 dichromat_2.0-0.1
[49] abind_1.4-8 codetools_0.2-20 spatstat.random_3.4-1
[52] miniUI_0.1.2 spatstat.explore_3.4-3 listenv_0.9.1
[55] lattice_0.22-7 tibble_3.3.0 plyr_1.8.9
[58] shiny_1.11.1 ROCR_1.0-11 Rtsne_0.17
[61] future_1.58.0 fastDummies_1.7.5 survival_3.8-3
[64] polyclip_1.10-7 fitdistrplus_1.2-4 pillar_1.11.0
[67] KernSmooth_2.23-26 plotly_4.11.0 generics_0.1.4
[70] RcppHNSW_0.6.0 ggplot2_3.5.2 scales_1.4.0
[73] globals_0.18.0 xtable_1.8-4 glue_1.8.0
[76] lazyeval_0.2.2 tools_4.5.1 data.table_1.17.8
[79] RSpectra_0.16-2 locfit_1.5-9.12 RANN_2.6.2
[82] dotCall64_1.2 cowplot_1.2.0 grid_4.5.1
[85] tidyr_1.3.1 colorspace_2.1-1 nlme_3.1-168
[88] patchwork_1.3.1 cli_3.6.5 spatstat.sparse_3.1-0
[91] spam_2.11-1 viridisLite_0.4.2 dplyr_1.1.4
[94] uwot_0.2.3 gtable_0.3.6 digest_0.6.37
[97] progressr_0.15.1 ggrepel_0.9.6 htmlwidgets_1.6.4
[100] farver_2.1.2 htmltools_0.5.8.1 lifecycle_1.0.4
[103] httr_1.4.7 statmod_1.5.0 mime_0.13
[106] bit64_4.6.0-1 MASS_7.3-65
>
I would appreciate any guidance you might be able to offer.
Thanks and best wishes,
Rich
Richard Friedman
Herbert Irving Comprehensive Cancer Center
Columbia University Irving Medical Center

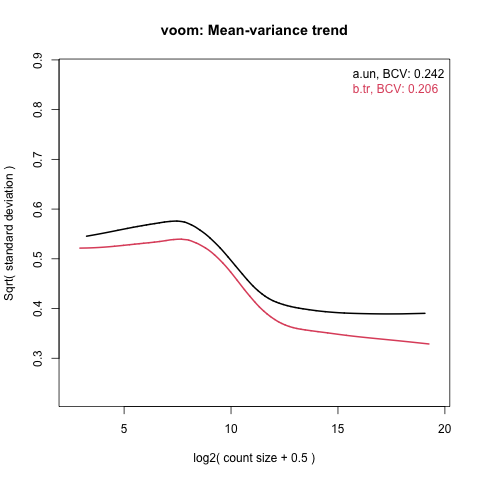

Thank you. I willleave as is and proceed.