We are trying to look at genes that are differentially expressed between three species, for two of those species we have data for 2 strains. The PCA shows that largest difference is between two species
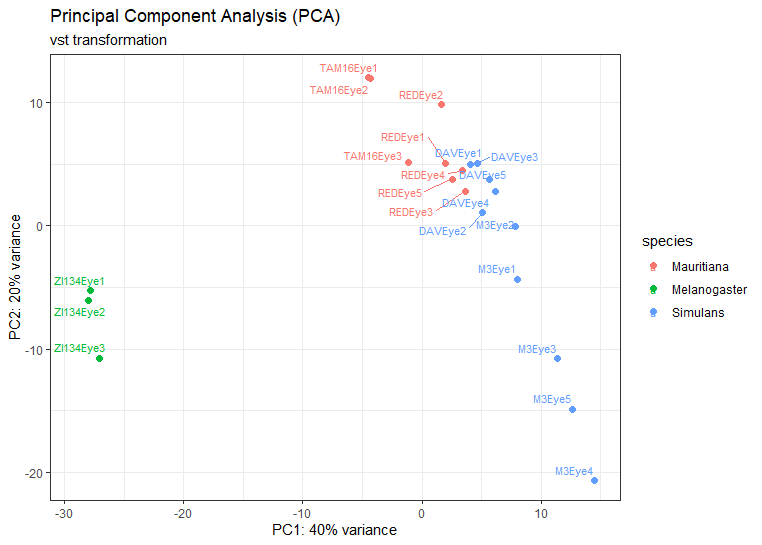
I wanted to know what the best approach would be to identify differentially expressed genes between species: to use a model comparing species (~species) or to do pairwise comparisons between the strains for the different species i.e (ZI134 vs M3 and ZI134 vs DAV) and then look at which genes overlap. Our fear is that by comparing species we might get genes that are differentially expressed between melanogaster and one of the strains within a species but not the other (missing intraspecific variation).

