Hi everyone,
I understand that when performing differential analysis between different groups using limma, it is better to include all samples and using contrasts rather than subsetting the groups to better estimate the variance.
However, I have a complex design where this is causing me an issue.
For context, I am performing differential methylation analysis between 3 groups. This is a reduced example dataset of my analysis with my study design (I have a larger dataset and observing the same issue described here) :
df = structure(list(Group = c("G1", "G2", "G1", "G2", "G2", "G1",
"G3", "G3", "G3", "G3", "G3", "G3", "G3"), V1 = c(36L, 36L, 26L,
26L, 34L, 34L, 40L, 29L, 34L, 31L, 34L, 35L, 25L), V2 = c("Covar1",
"Covar1", "Covar1", "Covar1", "Covar2", "Covar2", "Covar2", "Covar3",
"Covar1", "Covar2", "Covar2", "Covar2", "Covar1"), Subject = structure(c(3L,
3L, 6L, 6L, 7L, 7L, 1L, 2L, 4L, 5L, 8L, 9L, 10L), levels = c("1",
"2", "3", "4", "6", "8", "13", "16", "18", "23"), class = "factor")), row.names = c(NA,
-13L), class = "data.frame")
design <- model.matrix(~0+Group+V1+V2+Subject, data=df)
beta_test <- c(0.598403375449404, 0.635837157363606, 0.577366021553369, 0.614350852641186,
0.979187240034327, 0.964493441174273, 0.976979399329944, 0.976636922254339,
0.973782723774806, 0.979580729724208, 0.971783039226741, 0.0381216484812949,
0.647705918700804)
fit <- lmFit(beta_test, design)
contr <- makeContrasts(GroupG2 - GroupG3, levels = colnames(coef(fit)))
fit2 <- contrasts.fit(fit, contr)
fit2 <- eBayes(fit2)
degrees.freedom = fit2$df.residual - 1
DMPs=data.frame(effectsize=fit2$coefficients,SE=fit2$stdev.unscaled*fit2$sigma,Pval=fit2$p.value,degrees.freedom=degrees.freedom)
DMPs$probe=rownames(DMPs)
colnames(DMPs)=c("effectsize",'SE',"Pval","degrees.freedom","probe")
DMPs
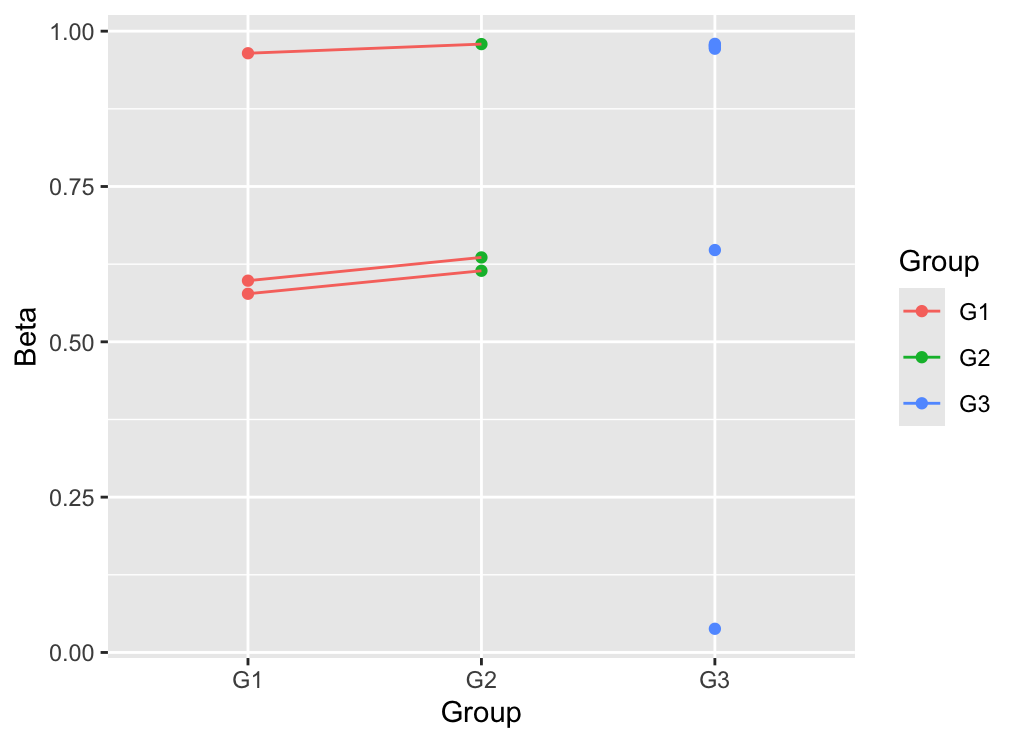
So I have 3 groups, with 3 covariates that I want to adjust in my model (V1,V2, Subject). For 2 groups (G1/G2), samples are paired (coming from the same individual) while the remaining group (G3) are samples from other individuals. I use contrasts to compare G1, G2 and G3 but when comparing G3 vs. G1 and G3 vs. G2, I have a problem with effect sizes. Indeed my data are beta values that ranges between 0 and 1 (I know it is not optimal here). However, the effect size announced by the G3 vs. G1 here is 1.13 which is not reliable.
When I remove the Subject covariate in the design, the effect size is 0.03 which is more logical when looking at the data.
I am sorry as my knowledge is limited in statistics and my question may be pointless, but why is there such a huge variation between both models while there is not a big effect of Subject according to the plot (dots that are connected are from the same subject)?
Is it ok to perform the analysis without the Subject for G3 vs. G1 and G3 vs. G2 but keeping the Subject for G2 vs. G1 and thus changing the design while comparing the groups ?
Thank you in advance for your help


Thank you very much for your time and assistance