Hello!
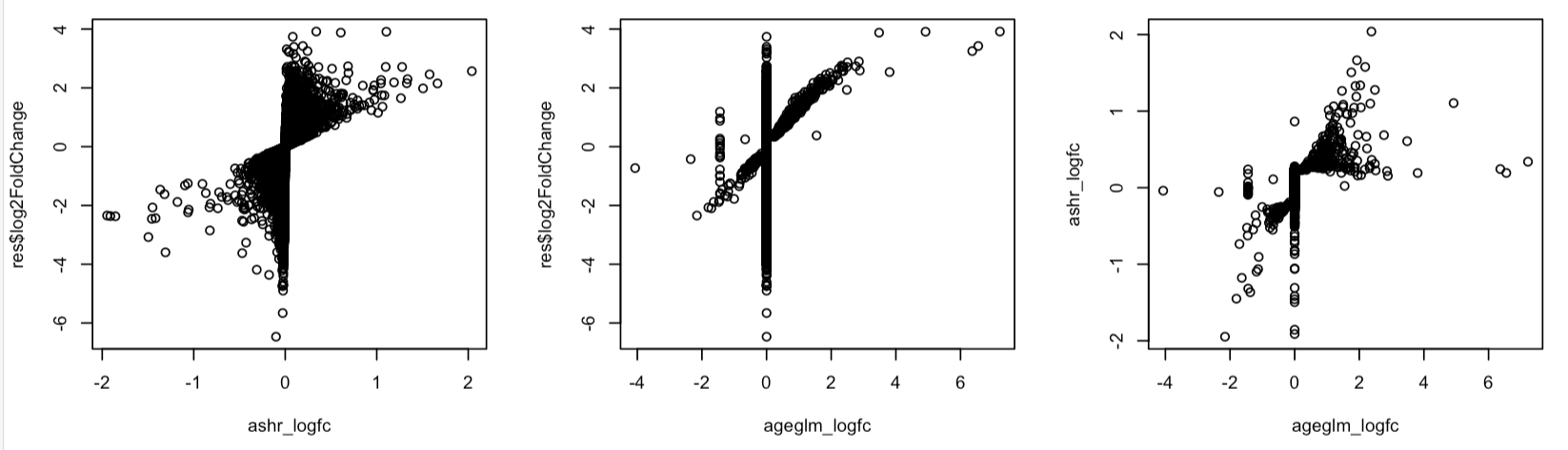
I'm analyzing pseudobulked single-cell RNA-seq data using DESeq2 and comparing apeglm vs ashr for log fold change shrinkage. Even in my largest cell type (comparing 8 vs 11 samples, 25-7771 cells per sample with median cell number of 835), I'm observing substantial differences between the two methods that significantly impact downstream analysis.
Key observations:
- apeglm appears much more aggressive in shrinkage, pushing many genes toward logFC = 0
- ashr maintains a gradient of shrinkage values and preserves more moderate effect sizes
- In downstream GSEA analysis, apeglm yields very few significant enrichments while ashr produces many biologically plausible pathway enrichments
Below I'm showing the correlation between original and shrinked logFCs:
Specific question:
For pseudobulked scRNA-seq data, are there methodological reasons to prefer one approach over the other? I'm particularly interested in:
- Whether the distributional assumptions of each method are better suited to the characteristics of pseudobulked data
- If the more aggressive shrinkage by apeglm might be overly conservative for this data type, potentially masking true biological signal
- How to objectively evaluate which approach is more appropriate when the biological interpretation seems more coherent with one method
I want to avoid confirmation bias in method selection - while ashr results align better with my biological expectations, I'm concerned this might influence my judgment. Are there principled ways to evaluate shrinkage method appropriateness beyond downstream biological plausibility?
Thank you!