Hello,
I'm wondering if there was a change in a dependency for the dispersion estimate calculation or how the best fit is decided. I reran some code to do analysis of ATAC-seq data and noticed that my results were very different than my initial run because of a change in the dispersion fit from parametric to local. The local fit has a much higher result for the median absolute residual than my initial run using the parametric fit, and given the same version of DESeq2 being used for both calculations I'm wondering if there is a dependency that got updated somewhere that is causing this change in how the fit is determined.
# Code used to generate DESeq2 output (same in both runs with different results d/t fitType)
# Load in the previously produced counts table
count_df <- ct
head(count_df)
# A549_KO_CTRL_BRep1 A549_KO_CTRL_BRep2 A549_KO_DP_BRep1 A549_KO_DP_BRep2 A549_WT_CTRL_BRep1 A549_WT_CTRL_BRep2 A549_WT_DP_BRep1 A549_WT_DP_BRep2
# chr1:180984-181655 4 10 29 76 2 2 35 47
# chr1:191406-191640 10 9 10 19 8 9 13 31
# chr1:267959-268180 39 47 20 39 28 43 21 48
# chr1:778408-779163 347 514 167 326 129 386 148 180
# chr1:827090-827791 92 157 45 121 21 95 46 63
# chr1:869715-870091 67 99 25 80 19 60 22 44
# Create a sample information data frame by separating the column names of the counts data frame
sample_info <- strsplit(colnames(count_df), split = "_")
sample_df <- do.call(rbind, sample_info)
colnames(sample_df) <- c("Cell", "Mut", "Treatment", "Rep")
sample_df <- as.data.frame(sample_df)
rownames(sample_df) <- paste0(sample_df$Cell, "_", sample_df$Mut, "_", sample_df$Treatment, "_", sample_df$Rep)
colnames(count_df) <- rownames(sample_df)
# Create a group variable that is a concatenation of the mutational status and treatment
sample_df$group <- paste(sample_df$Mut, sample_df$Treatment, sep = "_")
sample_df
# Cell Mut Treatment Rep group
# A549_KO_CTRL_BRep1 A549 KO CTRL BRep1 KO_CTRL
# A549_KO_CTRL_BRep2 A549 KO CTRL BRep2 KO_CTRL
# A549_KO_DP_BRep1 A549 KO DP BRep1 KO_DP
# A549_KO_DP_BRep2 A549 KO DP BRep2 KO_DP
# A549_WT_CTRL_BRep1 A549 WT CTRL BRep1 WT_CTRL
# A549_WT_CTRL_BRep2 A549 WT CTRL BRep2 WT_CTRL
# A549_WT_DP_BRep1 A549 WT DP BRep1 WT_DP
# A549_WT_DP_BRep2 A549 WT DP BRep2 WT_DP
# Create a dds object to run DESeq2
dds <- DESeqDataSetFromMatrix(countData = count_df,
colData = sample_df,
design = ~ Rep + group)
# Run DESeq2 and pull results for the same comparisons as RNAseq
dds1 <- DESeq(dds)
# estimating size factors
# estimating dispersions
# gene-wise dispersion estimates
# mean-dispersion relationship
# -- note: fitType='parametric', but the dispersion trend was not well captured by the
# function: y = a/x + b, and a local regression fit was automatically substituted.
# specify fitType='local' or 'mean' to avoid this message next time.
# final dispersion estimates
# fitting model and testing
# Compare the two dds objects
# Show that the original DDS object loaded (orig_dds) has the same data
identical(counts(orig_dds) %>% as.data.frame, counts(dds1) %>% as.data.frame)
# [1] TRUE
# Show the difference in the observed and prior variance of parametric and local fit
# Parametric fit
orig_dds@dispersionFunction
# function (q)
# coefs[1] + coefs[2]/q
# <bytecode: 0x565313622910>
# <environment: 0x5653135cd818>
# attr(,"coefficients")
# asymptDisp extraPois
# 0.01427565 2.07295375
# attr(,"fitType")
# [1] "parametric"
# attr(,"varLogDispEsts")
# [1] 0.5034352
# attr(,"dispPriorVar")
# [1] 0.25
# Local fit
dds1@dispersionFunction
# function (means)
# exp(predict(fit, data.frame(logMeans = log(means))))
# <bytecode: 0x565314280498>
# <environment: 0x56531474e398>
# attr(,"fitType")
# [1] "local"
# attr(,"varLogDispEsts")
# [1] 0.1674445
# attr(,"dispPriorVar")
# [1] 0.7607608
# Show that the median absolute residual of the initial parametric fit is lower than the local fit
local_MAR <- median(mcols(dds1)$dispGeneEst - mcols(dds1)$dispFit) %>% abs
local_MAR
#> [1] 3.160043
parametric_MAR <- local_residual <- median(mcols(orig_dds)$dispGeneEst - mcols(orig_dds)$dispFit) %>% abs
parametric_MAR
#> [1] 0.05677162
# Show the DESeq2 versions are the same between runs
dds1@metadata$version
#> [1] `1.44.0`
orig_dds@metadata$version
#> [1] `1.44.0`
# Plot the dispersion estimates
plotDispEsts(dds1, main = "Local Fit")
plotDispEsts(orig_dds, main = "Parametric Fit")
sessionInfo()
R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.5 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplotify_0.1.2 DiffBind_3.14.0 DESeq2_1.44.0 SummarizedExperiment_1.34.0 Biobase_2.64.0 MatrixGenerics_1.16.0 matrixStats_1.5.0
[8] plyranges_1.24.0 GenomicRanges_1.56.2 GenomeInfoDb_1.40.1 IRanges_2.38.1 S4Vectors_0.42.1 BiocGenerics_0.50.0 epitools_0.5-10.1
[15] eulerr_7.0.2 ggsankey_0.0.99999 ggpubr_0.6.1 svglite_2.2.1 ggrepel_0.9.6 ggh4x_0.3.1 enrichplot_1.24.4
[22] viridisLite_0.4.2 patchwork_1.3.2 plyr_1.8.9 lubridate_1.9.4 forcats_1.0.0 stringr_1.5.2 dplyr_1.1.4
[29] purrr_1.1.0 readr_2.1.5 tidyr_1.3.1 tibble_3.3.0 ggplot2_4.0.0 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] R.methodsS3_1.8.2 dichromat_2.0-0.1 vroom_1.6.5 goftest_1.2-3 Biostrings_2.72.1 vctrs_0.6.5 spatstat.random_3.4-1 digest_0.6.37
[9] png_0.1-8 mixsqp_0.3-54 deldir_2.0-4 parallelly_1.45.1 MASS_7.3-60.2 SQUAREM_2021.1 reshape2_1.4.4 httpuv_1.6.16
[17] qvalue_2.36.0 withr_3.0.2 xfun_0.53 amap_0.8-20 ggfun_0.2.0 survival_3.6-4 memoise_2.0.1 systemfonts_1.2.3
[25] tidytree_0.4.6 zoo_1.8-14 gtools_3.9.5 pbapply_1.7-4 R.oo_1.27.1 Formula_1.2-5 KEGGREST_1.44.1 promises_1.3.3
[33] httr_1.4.7 GreyListChIP_1.36.0 rstatix_0.7.2 restfulr_0.0.16 globals_0.18.0 fitdistrplus_1.2-4 ashr_2.2-63 rstudioapi_0.17.1
[41] UCSC.utils_1.0.0 miniUI_0.1.2 generics_0.1.4 DOSE_3.30.5 curl_7.0.0 zlibbioc_1.50.0 ggraph_2.2.2 polyclip_1.10-7
[49] GenomeInfoDbData_1.2.12 SparseArray_1.4.8 xtable_1.8-4 evaluate_1.0.5 S4Arrays_1.4.1 systemPipeR_2.10.0 hms_1.1.3 irlba_2.3.5.1
[57] colorspace_2.1-1 ROCR_1.0-11 reticulate_1.43.0 spatstat.data_3.1-8 magrittr_2.0.3 lmtest_0.9-40 later_1.4.4 viridis_0.6.5
[65] ggtree_3.12.0 lattice_0.22-6 spatstat.geom_3.5-0 future.apply_1.20.0 scattermore_1.2 XML_3.99-0.19 shadowtext_0.1.6 cowplot_1.2.0
[73] RcppAnnoy_0.0.22 pillar_1.11.0 nlme_3.1-164 pwalign_1.0.0 caTools_1.18.3 compiler_4.4.1 RSpectra_0.16-2 stringi_1.8.7
[81] tensor_1.5.1 GenomicAlignments_1.40.0 crayon_1.5.3 abind_1.4-8 BiocIO_1.14.0 truncnorm_1.0-9 gridGraphics_0.5-1 emdbook_1.3.14
[89] locfit_1.5-9.12 sp_2.2-0 graphlayouts_1.2.2 bit_4.6.0 fastmatch_1.1-6 codetools_0.2-20 textshaping_1.0.3 plotly_4.11.0
[97] mime_0.13 splines_4.4.1 Rcpp_1.1.0 fastDummies_1.7.5 utilsFanc_0.1.0 interp_1.1-6 knitr_1.50 blob_1.2.4
[105] apeglm_1.26.1 fs_1.6.6 listenv_0.9.1 ggsignif_0.6.4 Matrix_1.7-0 statmod_1.5.0 tzdb_0.5.0 tweenr_2.0.3
[113] pkgconfig_2.0.3 tools_4.4.1 cachem_1.1.0 RSQLite_2.4.3 DBI_1.2.3 numDeriv_2016.8-1.1 fastmap_1.2.0 rmarkdown_2.29
[121] scales_1.4.0 grid_4.4.1 ica_1.0-3 Seurat_5.3.0 Rsamtools_2.20.0 broom_1.0.9 coda_0.19-4.1 dotCall64_1.2
[129] carData_3.0-5 RANN_2.6.2 farver_2.1.2 tidygraph_1.3.1 scatterpie_0.2.5 yaml_2.3.10 latticeExtra_0.6-31 rtracklayer_1.64.0
[137] cli_3.6.5 lifecycle_1.0.4 uwot_0.2.3 mvtnorm_1.3-3 backports_1.5.0 BiocParallel_1.38.0 timechange_0.3.0 gtable_0.3.6
[145] rjson_0.2.23 ggridges_0.5.7 progressr_0.15.1 parallel_4.4.1 ape_5.8-1 limma_3.60.6 jsonlite_2.0.0 RcppHNSW_0.6.0
[153] bitops_1.0-9 bit64_4.6.0-1 Rtsne_0.17 yulab.utils_0.2.1 spatstat.utils_3.1-5 SeuratObject_5.2.0 bdsmatrix_1.3-7 GOSemSim_2.30.2
[161] spatstat.univar_3.1-4 R.utils_2.13.0 lazyeval_0.2.2 shiny_1.11.1 htmltools_0.5.8.1 GO.db_3.19.1 sctransform_0.4.2 rappdirs_0.3.3
[169] glue_1.8.0 spam_2.11-1 httr2_1.2.1 XVector_0.44.0 RCurl_1.98-1.17 treeio_1.28.0 BSgenome_1.72.0 jpeg_0.1-11
[177] gridExtra_2.3 invgamma_1.2 igraph_2.1.4 R6_2.6.1 ggiraph_0.9.0 gplots_3.2.0 cluster_2.1.6 bbmle_1.0.25.1
[185] aplot_0.2.8 DelayedArray_0.30.1 tidyselect_1.2.1 ggforce_0.5.0 car_3.1-3 AnnotationDbi_1.66.0 future_1.67.0 KernSmooth_2.23-24
[193] S7_0.2.0 data.table_1.17.8 htmlwidgets_1.6.4 fgsea_1.30.0 RColorBrewer_1.1-3 hwriter_1.3.2.1 rlang_1.1.6 spatstat.sparse_3.1-0
[201] spatstat.explore_3.5-2 uuid_1.2-1 ShortRead_1.62.0
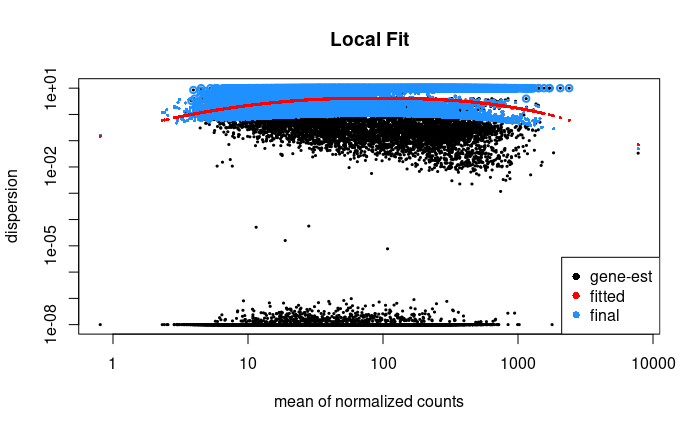

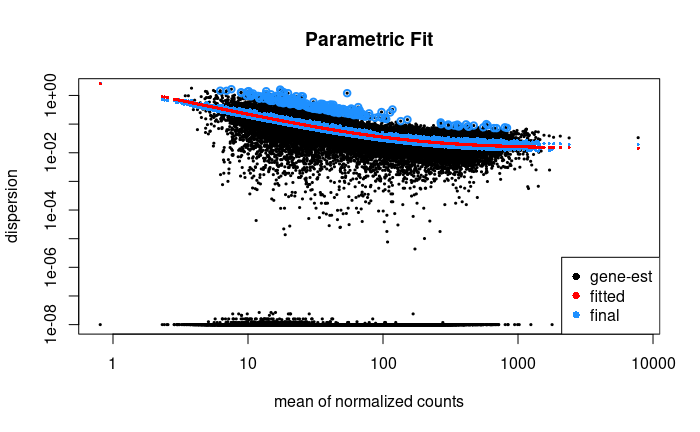

FWIW: This seems to be due to architectural differences between two compute nodes I was running on, specifically with the DESeq2:::fitNbinomGLMs() function. My guess is that differences in floating point calculations between different CPU architecture (but the same BLAS/LAPACK installation) led to differences because of columns with very small values in the input matrix, but I'm not certain. Running on nodes with the same CPU generation produced identical results, whereas newer nodes produced different results (but intra-generationally consistent).