Hi Bioconductor,
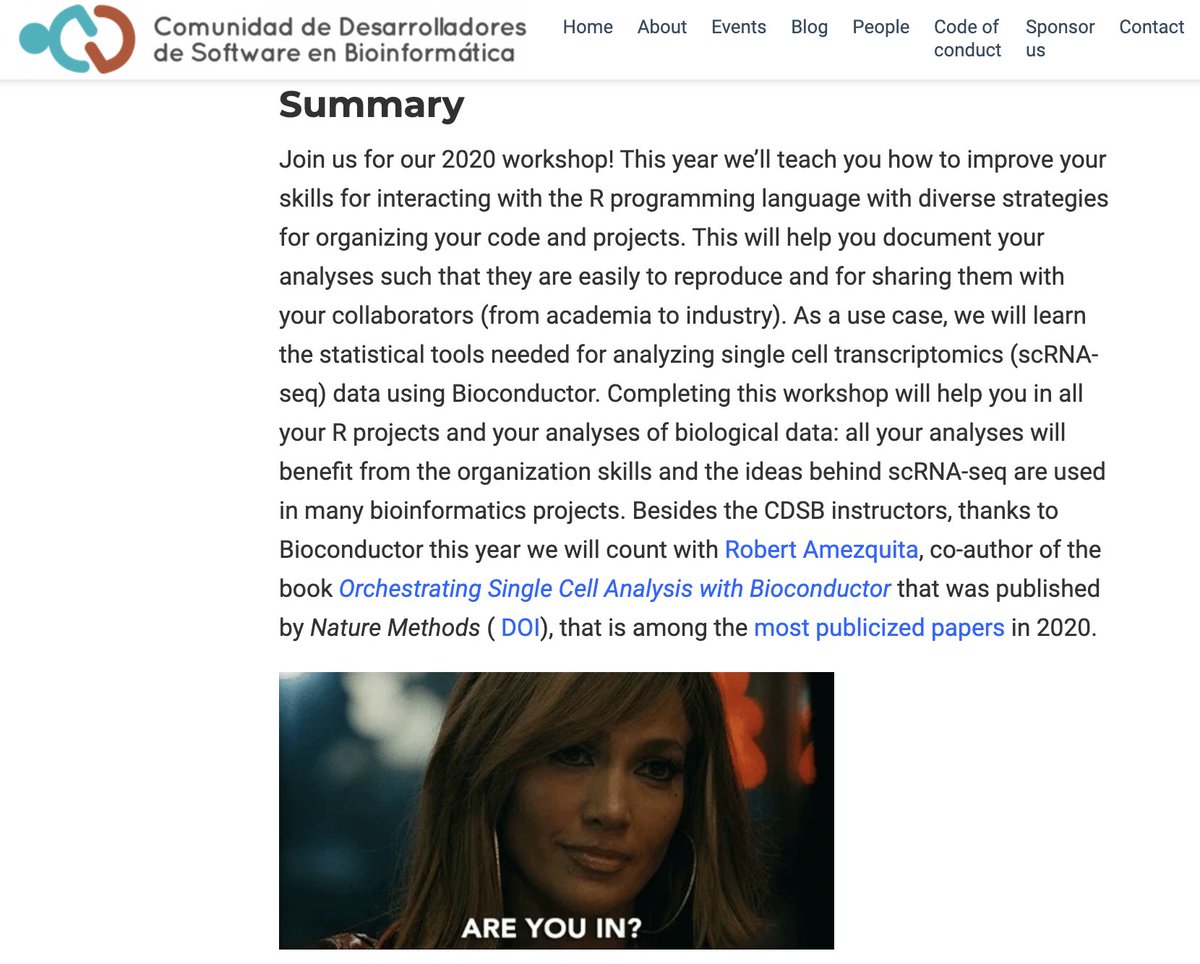
It's my pleasure to announce the CDSB Workshop 2020: Building workflows with RStudio and Bioconductor for single cell RNA-seq analysis. It will be held in Cuernavaca, Morelos, Mexico from August 3rd to the 7th, 2020. We are also grateful for the support from Bioconductor Foundation in making this workshop a reality.
If you know anyone that might be interested, please help us share the news. We are also looking for sponsors if you know any. We wrote the answer to why should you support us? at https://comunidadbioinfo.github.io/niveles-de-patrocinio/ (also available in Spanish as is all the website). Note that the workshop will be taught in English only as we have a guest instructor.
Here's the Facebook event page https://www.facebook.com/events/921536534948636/ and here's the announcement from Twitter:
Do you want to learn how to organize your R code 👨🏽💻 and analyze 👩🏽💻single cell RNA sequencing data with @Bioconductor?
— ComunidadBioInfo (@CDSBMexico) March 6, 2020
Save the date! See you August 3-7🗓️
👩🏽🏫👨🏽🏫 @AleMedinaRivera @fellgernon @josschavezf1 @robamezquita @TeresaOM#rstatES #scRNAseq ¡🙏🏽 RT!https://t.co/Nw5RKS6BFE pic.twitter.com/2Op9AvYcrL
English version (en inglés)
Do you want to learn how to organize your R code 👨🏽💻 and analyze 👩🏽💻single cell RNA sequencing data with @Bioconductor?
Save the date! See you August 3-7 🗓
Instructors 👩🏽🏫👨🏽🏫 : @AleMedinaRivera @fellgernon @josschavezf1 @robamezquita @TeresaOM
#rstats #scRNAseq ¡🙏🏽 please share!
https://comunidadbioinfo.github.io/post/cdsb2020-building-workflows-with-rstudio-and-scrnaseq-with-bioconductor/
Spanish version (Español)
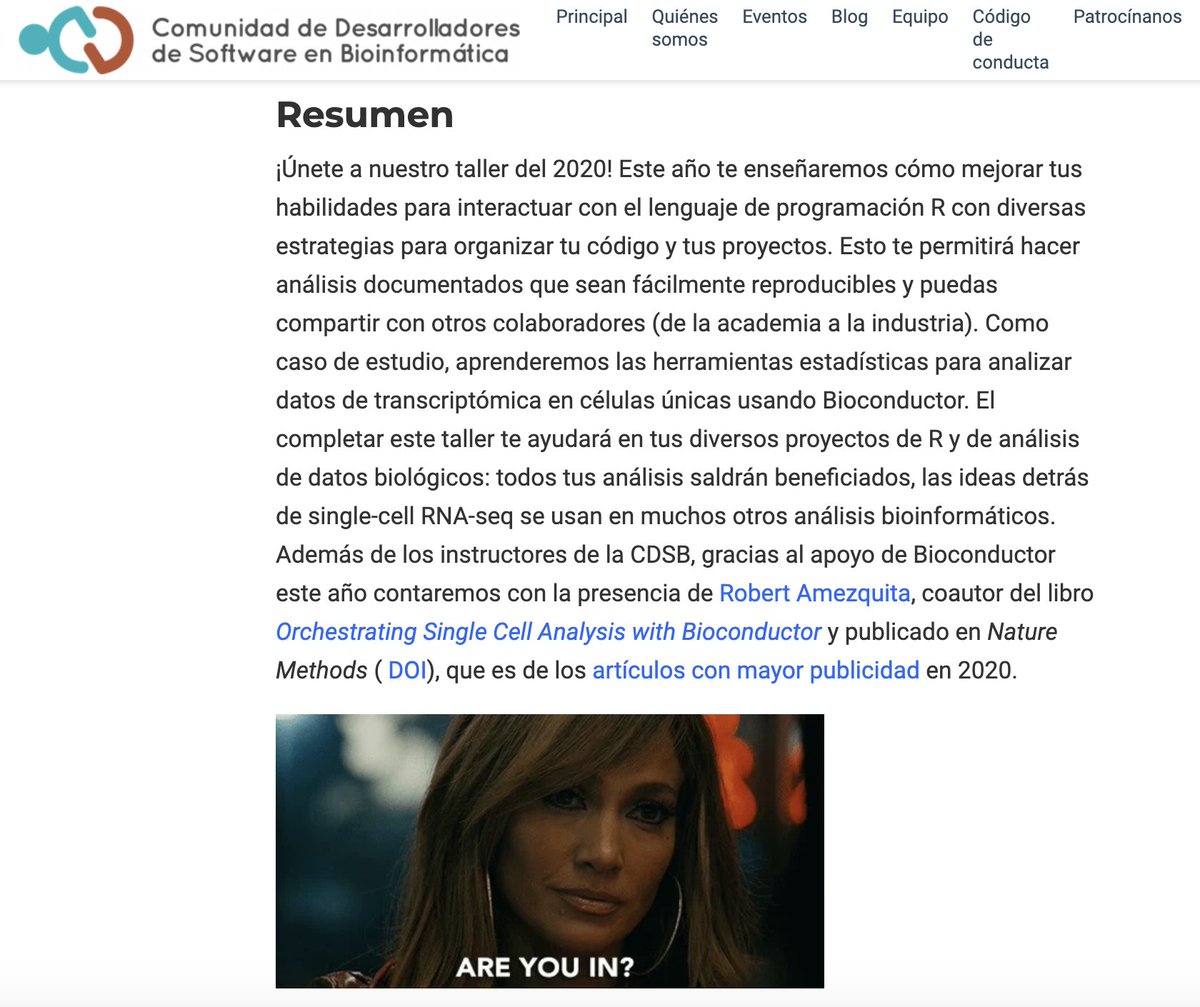
¿Quieres aprender a organizar tu código de R 👩🏽💻 y a analizar datos 👨🏽💻de secuenciación de células únicas con @Bioconductor?
¡Te esperamos 3-7 de agosto! Aparta la fecha
Instructores 👩🏽🏫👨🏽🏫 : @AleMedinaRivera @fellgernon @josschavezf1 @robamezquita @TeresaOM
#rstatsES #scRNAseq ¡🙏🏽 favor de compartir!
https://comunidadbioinfo.github.io/es/post/cdsb2020-building-workflows-with-rstudio-and-scrnaseq-with-bioconductor/