Hi, I'm trying to create a dataframe that contains the attributes (interpro_description) of each of the correspoding ensemblID found within a column (ensemblID) of another dataframe (joinedData). I've followed the vignette for biomaRt accordingly, but I keep getting the same error. Anyone has any idea how to resolve the error? The code was working prior, but it seems to stop working somehow.
mart <- useMart(biomart= "ENSEMBL_MART_MOUSE")
mart <- useDataset(dataset = 'mmc57bl6nj_gene_ensembl', mart=mart)
getBM(attributes = c("interpro_description"),
filters= "arrayexpress",
values= joinedData$ensemblID,
mart=mart)
Error output: Error in getNodeSet(html, path = "//div[@class='plain-box float-right archive-box']")[[1]] : subscript out of bounds
sessionInfo()
R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252 LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 biomaRt_2.46.3
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 cellranger_1.1.0 pillar_1.4.7
[4] compiler_4.0.3 BiocManager_1.30.10 dbplyr_2.1.0
[7] prettyunits_1.1.1 tools_4.0.3 progress_1.2.2
[10] bit_4.0.4 tibble_3.0.6 RSQLite_2.2.3
[13] memoise_2.0.0 BiocFileCache_1.14.0 lifecycle_1.0.0
[16] pkgconfig_2.0.3 rlang_0.4.10 DBI_1.1.1
[19] curl_4.3 parallel_4.0.3 fastmap_1.1.0
[22] dplyr_1.0.4 stringr_1.4.0 httr_1.4.2
[25] xml2_1.3.2 rappdirs_0.3.3 generics_0.1.0
[28] S4Vectors_0.28.1 vctrs_0.3.6 askpass_1.1
[31] IRanges_2.24.1 hms_1.0.0 tidyselect_1.1.0
[34] stats4_4.0.3 bit64_4.0.5 glue_1.4.2
[37] Biobase_2.50.0 R6_2.5.0 AnnotationDbi_1.52.0
[40] XML_3.99-0.5 purrr_0.3.4 blob_1.2.1
[43] magrittr_2.0.1 ellipsis_0.3.1 BiocGenerics_0.36.0
[46] assertthat_0.2.1 stringi_1.5.3 openssl_1.4.3
[49] cachem_1.0.3 crayon_1.4.1

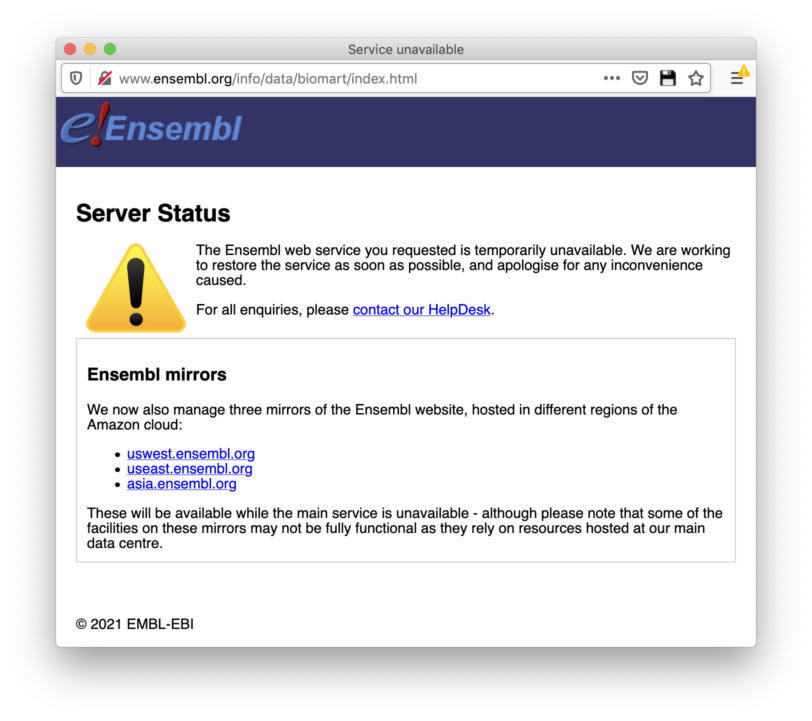

A call to this function listEnsemblArchives() returns the same error in R.
But I am able to visit this link (https://www.ensembl.org/info/website/archives/index.html) from my browser
I haven't done anything between today and yesterday, except restarting R and my computer and the problem seems to be resolved on its own. Haven't had a clue as to why the problem occurred in the first place or what change for it to be resolved.
Regardless, thanks @ Mike Smith for reaching out to assist.
Thanks for reporting back that it's now working. Hopefully it was just temporary issue with the Ensembl site, but thank you for reporting the issue. I'll try to add some better error handling to biomaRt to catch this problem if it occurs again and offer some guidance to users.
Hello, I am confronted with the exact same error and can't seem to get the getBM function to work despite having restarted R. Is there a way to bypass the internal call to listEnsemblArchives()?
I am also having the same issue with this error when I run the getBM() function and when I run the function listEnsemblArchives().
When I go to the Website (https://www.ensembl.org/info/website/archives/index.html) I get the following error: "The Ensembl web service you requested is temporarily unavailable. We are working to restore the service as soon as possible, and apologise for any inconvenience caused."
I am getting this same error right now.
I am also getting the same error. I just tried now.