Entering edit mode
Eslam
•
0
@eslam-24990
Last seen 4.9 years ago
I'm trying to install wanted R packages by using the following code
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")
BiocManager::install("org.Hs.eg.db") but the installation failed

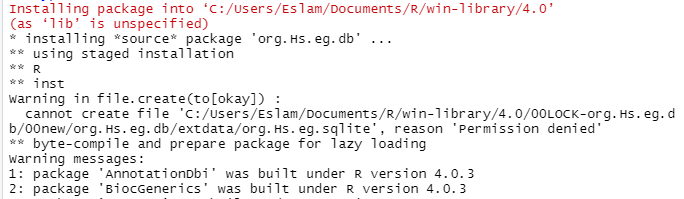

I used your code and went to the dir but did not find 00LOCK file and still facing the same problem
and try to manually construct new file in this dir with the name of (org.Hs.eg.db) or (00LOCK-org.Hs.eg.db) and restart and install again but still facing same problem also the constructed file in this dir was removed even save it opened during the installation.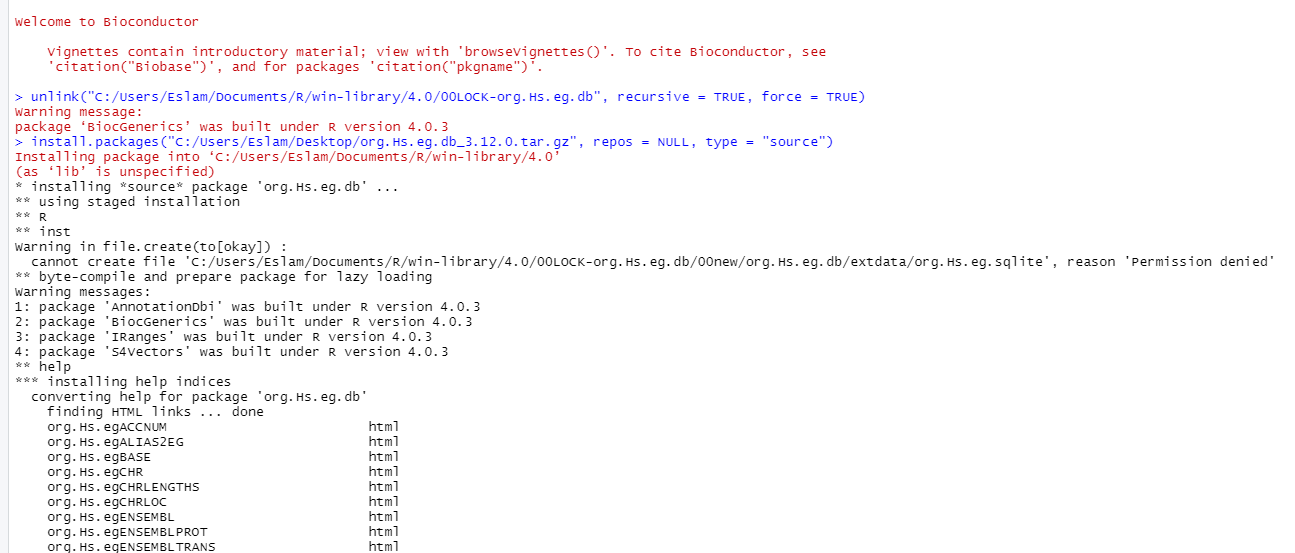
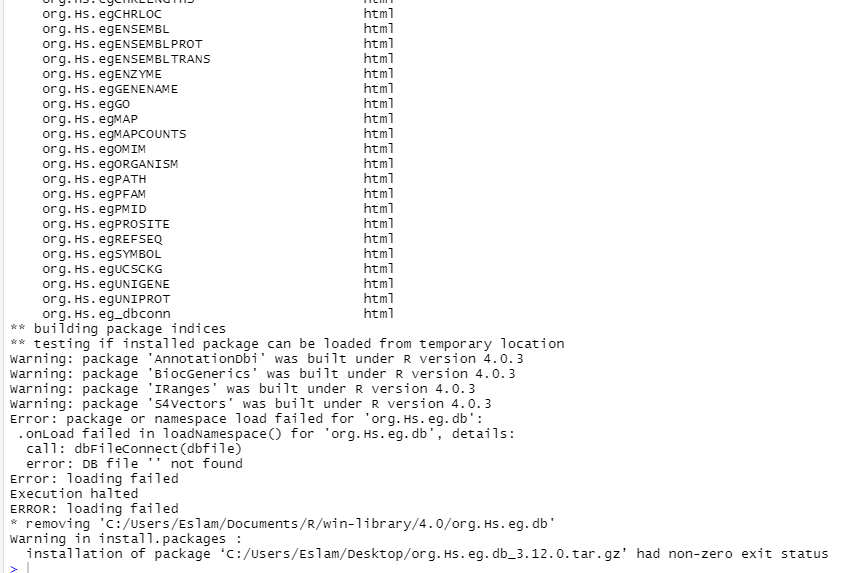
Linking to pictures of your console output is suboptimal. Instead, copy/paste the output as text and follow the example presented here to format it correctly.
I'm, assuming you can install other packages to that library directory? How about org.Mm.eg.db?
As part of the install process, R creates a temporary directory called 00LOCK-org.Hs.eg.db to do a temporary install of the package. You shouldn't ever have a directory with that name, except for the time when the package is being actively installed. The fact that you have an issue with permissions for just one file makes me think that you actually do have that directory, and there is already an org.Hs.eg.sqlite file there. Which is why you need to delete it. And you shouldn't be creating the directory for R, it will make the directory itself.
Thank you; I will take it into consideration the next time, the same problem appeared with org.Mm.eg.db but, I followed the steps in youtube video () as follow: 1- run R.stedio as administrator. 2- change the directory of the installation rather than the default one. 3- manually install the package with the specific installation problem. I did that and already the two packages have been installed.
thank you very much