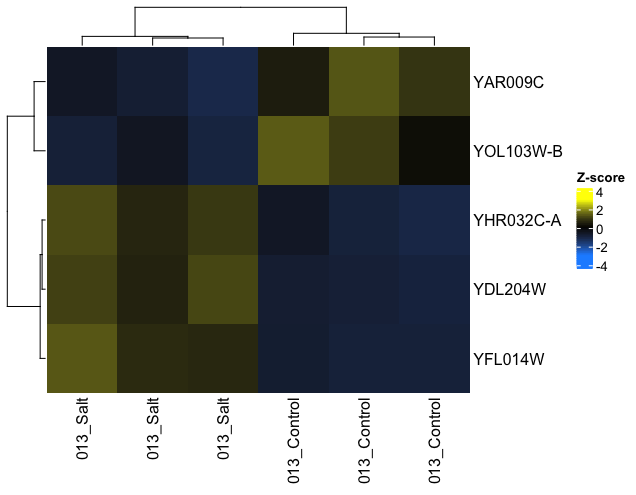
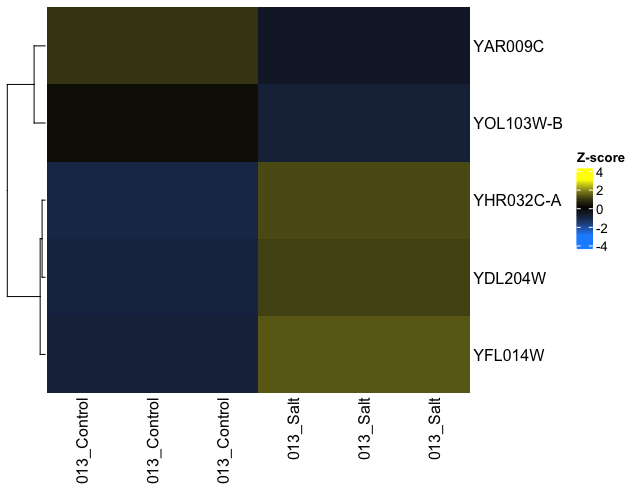
I'm using ComplexHeatmap to plot z-scaled data. I want to set the order of my samples (columns), which can be done by setting column_order=. However, when I use this function, the z-score values that are plotted change - it looks like the average value for the samples within a group are now plotted.
Here's a simplified example using values from 5 genes. Values for my R variables are in code chunks at the bottom.
library(DESeq2)
library(ComplexHeatmap)
library(circlize)
rlogTop25 <- (assay(rld)[sigTop25,]
z.TopLFC <- t(scale(t(rlogTop25), center=TRUE, scale=TRUE))
#Correctly Plots
Heatmap(z.TopLFC,
col = colorRamp2(c(-3, 0, 3), c('dodgerblue', 'black', 'yellow')),
heatmap_legend_param = list (
title = "Z-score")
)
#Plots averaged(?) Z-values for group of samples
Heatmap(z.TopLFC,
col = colorRamp2(c(-3, 0, 3), c('dodgerblue', 'black', 'yellow')),
column_order=sort(colnames(z.TopLFC)),
heatmap_legend_param = list (
title = "Z-score")
)
> rlogTop25
013_Control 013_Salt 013_Control 013_Salt 013_Control 013_Salt
YFL014W 6.347657 8.132517 6.462191 7.591956 6.360166 7.637381
YAR009C 10.166573 8.480758 9.699538 7.838010 10.682334 8.217700
YDL204W 7.560660 9.085627 7.693605 9.150839 7.633052 8.736366
YHR032C-A 5.822386 6.743593 6.025686 6.639762 5.880813 6.519343
YOL103W-B 8.929396 8.151790 9.847492 8.064004 9.552077 8.415369
> z.TopLFC
013_Control 013_Salt 013_Control 013_Salt 013_Control 013_Salt
YFL014W -0.9385917 1.3222492 -0.7935150 0.6375331 -0.9227476 0.6950720
YAR009C 0.8505378 -0.6040332 0.4475662 -1.1586147 1.2955512 -0.8310072
YDL204W -0.9856466 1.0201568 -0.8107826 1.1059313 -0.8904295 0.5607705
YHR032C-A -1.1002669 1.1544011 -0.6026881 0.9002739 -0.9572662 0.6055462
YOL103W-B 0.1376152 -0.9042738 1.3677420 -1.0218953 0.9719247 -0.5511128
attr(,"scaled:center")
YFL014W YAR009C YDL204W YHR032C-A YOL103W-B
7.088644 9.180819 8.310025 6.271931 8.826688
attr(,"scaled:scale")
YFL014W YAR009C YDL204W YHR032C-A YOL103W-B
0.7894670 1.1589778 0.7602771 0.4085777 0.7463425
> sort(colnames(z.TopLFC))
[1] "013_Control" "013_Control" "013_Control" "013_Salt" "013_Salt" "013_Salt"