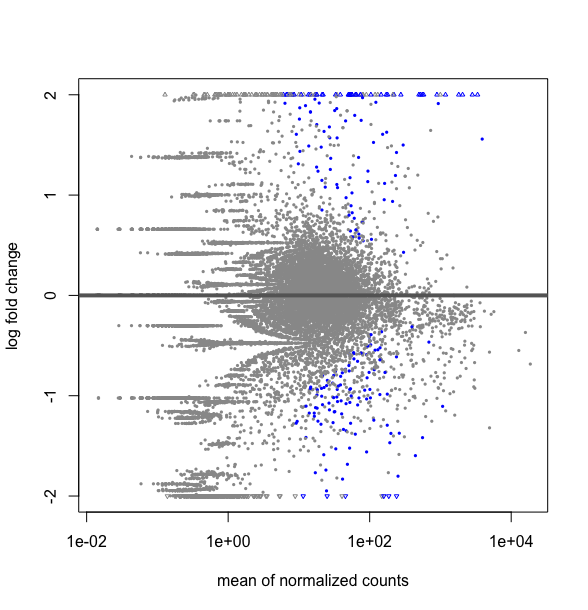
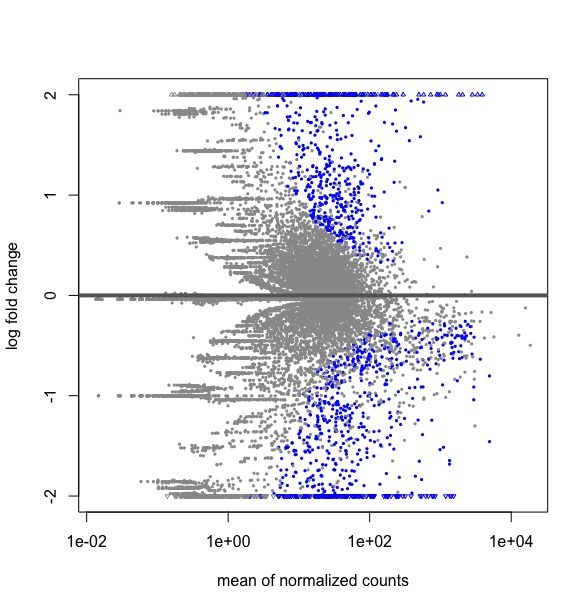
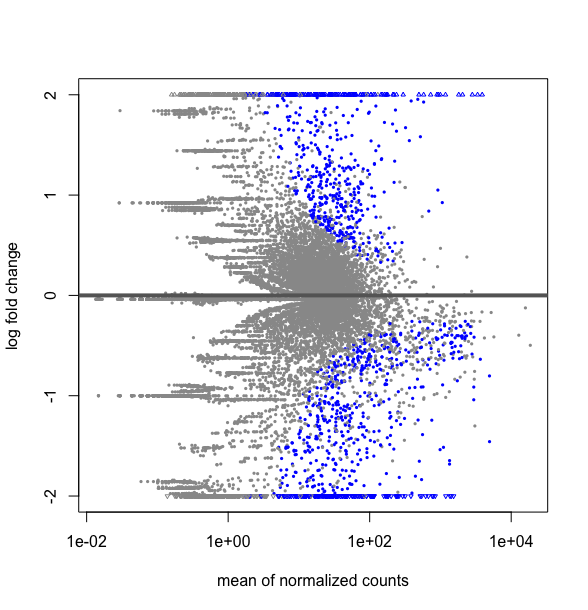
What is the difference between this two R codes, DESMatrix.dse being a DESeqDataSetFromMatrix with multiple 64 conditions including replicates:
plotMA(results(DESMatrix.dse, contrast=c("condition","treatedC","untreated")), ylim=c(-2,2))
# it gives the following plot :
and creating a sub matrix with only my two conditions
deseq2NI18vsUn.dse <- DESMatrix.dse[ , DESMatrix.dse@colData@listData$condition %in% c("treatedC","untreated" ]
deseq2NI18vsUn.res$condition
gives as expected: 1 untreated treatedC untreated treatedC [5] treatedC treatedC untreated
#then extracting the results to do the plot
res <- results(deseq2NI18vsUn.dse)
plotMA(res, ylim=c(-2,2))
This strangly looks equivalent to a plot over all the data obtained by:
plotMA(DESMatrix.dse, ylim=c(-2,2))
Thus is my submatrix deseq2NI18vsUn.dse really a subset of DESMatrix.dse ? If one can explain me why I do not get the same thing as I expected. Hope, I have given all the elements to understand the situation. Thanks for taking time to look to this.
Here are my R conditions:
sessionInfo( )
R version 4.0.4 (2021-02-15)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Mojave 10.14.6
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] fr_FR.UTF-8/fr_FR.UTF-8/fr_FR.UTF-8/C/fr_FR.UTF-8/fr_FR.UTF-8
attached base packages:
[1] stats4 parallel stats graphics grDevices utils datasets methods base
other attached packages:
[1] apeglm_1.12.0 kableExtra_1.3.4 org.Dm.eg.db_3.12.0 GOstats_2.56.0
[5] graph_1.68.0 Category_2.56.0 Matrix_1.3-2 AnnotationDbi_1.52.0
[9] ReportingTools_2.30.2 knitr_1.32 RColorBrewer_1.1-2 gplots_3.1.1
[13] ggplot2_3.3.3 dplyr_1.0.5 DESeq2_1.30.1 SummarizedExperiment_1.20.0
[17] MatrixGenerics_1.2.1 matrixStats_0.58.0 GenomicRanges_1.42.0 GenomeInfoDb_1.26.7
[21] IRanges_2.24.1 S4Vectors_0.28.1 Biobase_2.50.0 BiocGenerics_0.36.0
loaded via a namespace (and not attached):
[1] backports_1.2.1 Hmisc_4.5-0 systemfonts_1.0.1 BiocFileCache_1.14.0
[5] plyr_1.8.6 lazyeval_0.2.2 GSEABase_1.52.1 splines_4.0.4
[9] BiocParallel_1.24.1 digest_0.6.27 ensembldb_2.14.0 htmltools_0.5.1.1
[13] GO.db_3.12.1 fansi_0.4.2 magrittr_2.0.1 checkmate_2.0.0
[17] memoise_2.0.0 BSgenome_1.58.0 cluster_2.1.1 limma_3.46.0
[21] Biostrings_2.58.0 annotate_1.68.0 R.utils_2.10.1 svglite_2.0.0
[25] ggbio_1.38.0 bdsmatrix_1.3-4 askpass_1.1 prettyunits_1.1.1
[29] jpeg_0.1-8.1 colorspace_2.0-0 rvest_1.0.0 blob_1.2.1
[33] rappdirs_0.3.3 xfun_0.22 jsonlite_1.7.2 crayon_1.4.1
[37] RCurl_1.98-1.3 genefilter_1.72.1 survival_3.2-10 VariantAnnotation_1.36.0
[41] glue_1.4.2 gtable_0.3.0 zlibbioc_1.36.0 XVector_0.30.0
[45] webshot_0.5.2 DelayedArray_0.16.3 Rgraphviz_2.34.0 scales_1.1.1
[49] mvtnorm_1.1-1 DBI_1.1.1 GGally_2.1.1 edgeR_3.32.1
[53] Rcpp_1.0.6 emdbook_1.3.12 viridisLite_0.4.0 xtable_1.8-4
[57] progress_1.2.2 htmlTable_2.1.0 foreign_0.8-81 bit_4.0.4
[61] OrganismDbi_1.32.0 Formula_1.2-4 AnnotationForge_1.32.0 htmlwidgets_1.5.3
[65] httr_1.4.2 ellipsis_0.3.1 pkgconfig_2.0.3 reshape_0.8.8
[69] XML_3.99-0.6 R.methodsS3_1.8.1 sass_0.3.1 nnet_7.3-15
[73] dbplyr_2.1.1 locfit_1.5-9.4 utf8_1.2.1 tidyselect_1.1.0
[77] rlang_0.4.10 reshape2_1.4.4 munsell_0.5.0 tools_4.0.4
[81] cachem_1.0.4 generics_0.1.0 RSQLite_2.2.6 evaluate_0.14
[85] stringr_1.4.0 fastmap_1.1.0 yaml_2.2.1 bit64_4.0.5
[89] caTools_1.18.2 purrr_0.3.4 AnnotationFilter_1.14.0 RBGL_1.66.0
[93] R.oo_1.24.0 xml2_1.3.2 biomaRt_2.46.3 compiler_4.0.4
[97] rstudioapi_0.13 curl_4.3 png_0.1-7 PFAM.db_3.12.0
[101] tibble_3.1.0 geneplotter_1.68.0 bslib_0.2.4 stringi_1.5.3
[105] highr_0.8 GenomicFeatures_1.42.3 lattice_0.20-41 ProtGenerics_1.22.0
[109] vctrs_0.3.7 pillar_1.6.0 lifecycle_1.0.0 BiocManager_1.30.12
[113] jquerylib_0.1.3 data.table_1.14.0 bitops_1.0-6 rtracklayer_1.50.0
[117] hwriter_1.3.2 R6_2.5.0 latticeExtra_0.6-29 KernSmooth_2.23-18
[121] gridExtra_2.3 dichromat_2.0-0 MASS_7.3-53.1 gtools_3.8.2
[125] assertthat_0.2.1 openssl_1.4.3 withr_2.4.1 GenomicAlignments_1.26.0
[129] Rsamtools_2.6.0 GenomeInfoDbData_1.2.4 hms_1.0.0 grid_4.0.4
[133] rpart_4.1-15 coda_0.19-4 rmarkdown_2.7 biovizBase_1.38.0
[137] bbmle_1.0.23.1 numDeriv_2016.8-1.1 base64enc_0.1-3