Hello, I'm working with Arraystar lncRNA microarray for the first time. I tried to normalize the raw files (.txt) using the following script, which I used to normalize different other Agilent chip versions previously. Can anyone please tell me is this ok for the Arraystar chip also? Unfortunately, I'm not able to get the normalized values similar to those provided by the company. The company performed quantile normalization of the raw data using GeneSpring GX. Here is my script for quantile normalization:
library(limma)
targets <- readTargets("./targets.txt")
x <- read.maimages(targets, path="./Raw Data Files/", source="agilent",green.only=TRUE)
y <- backgroundCorrect(x, method="normexp", offset=16)
y <- normalizeBetweenArrays(y, method="quantile")
View(y$E)
y$E<-log2(y$E)
E = new("MAList", list(targets=y$targets, genes=y$genes, source=y$source, M=y$E, A=y$E))
E.avg <- avereps(E, ID=E$genes$ProbeName)


Hello, Sorry for a bit delayed reply. I got the idea to log2 transform the expression values from the following script: https://gist.github.com/brentp/1152860. In this script if you look at the line number 28, there is a log2 transformation step. But I also tried what you suggested i.e. analyzing the data skiping log2 transformation step. Then I saw that my boxplot is similar to the boxplot of the company normalized data, but the values are still not same. Hence I am getting different fold changes. Here the top image is the boxplot provided by the company, and the bottom image is the boxplot generated by me after skipping log2 transformation step.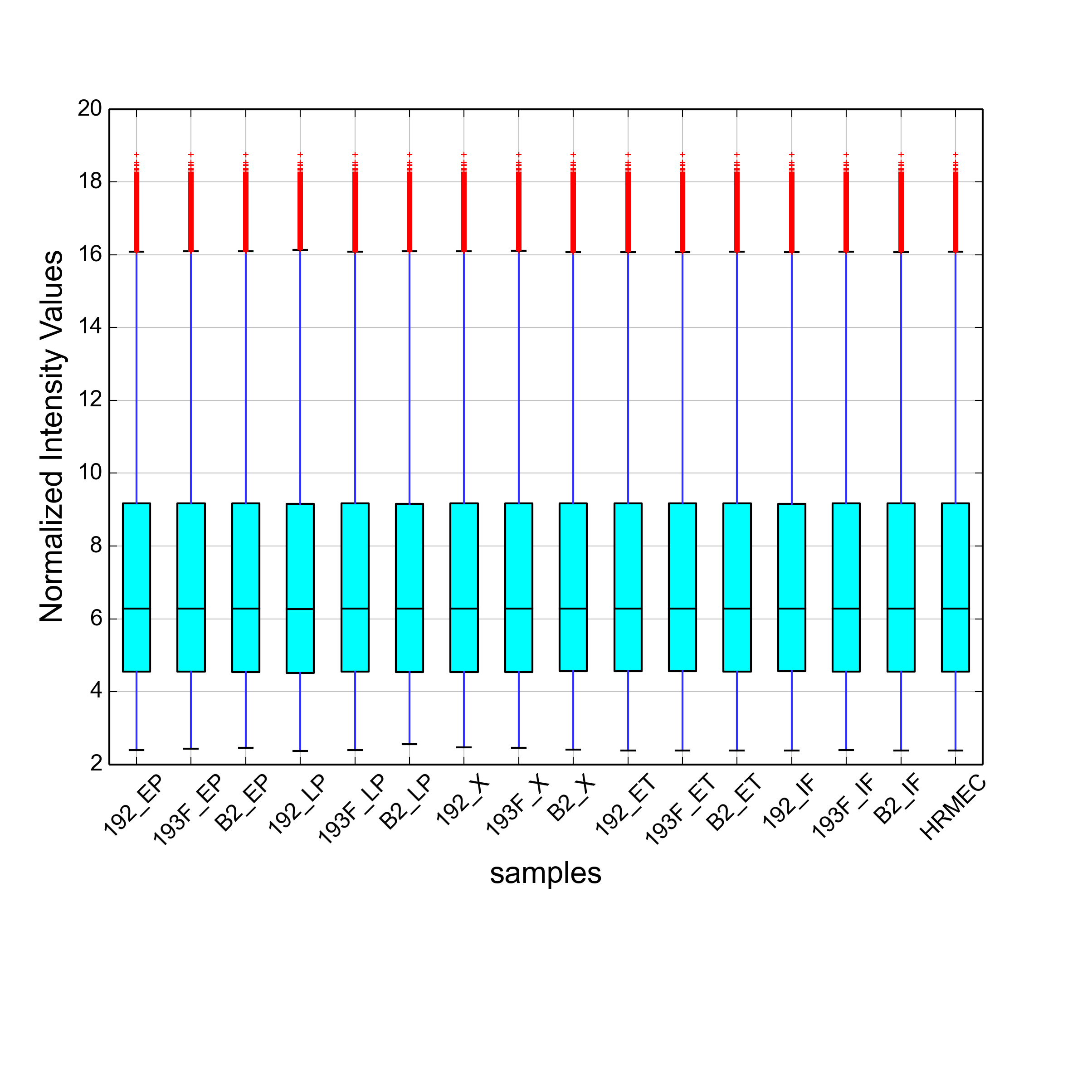
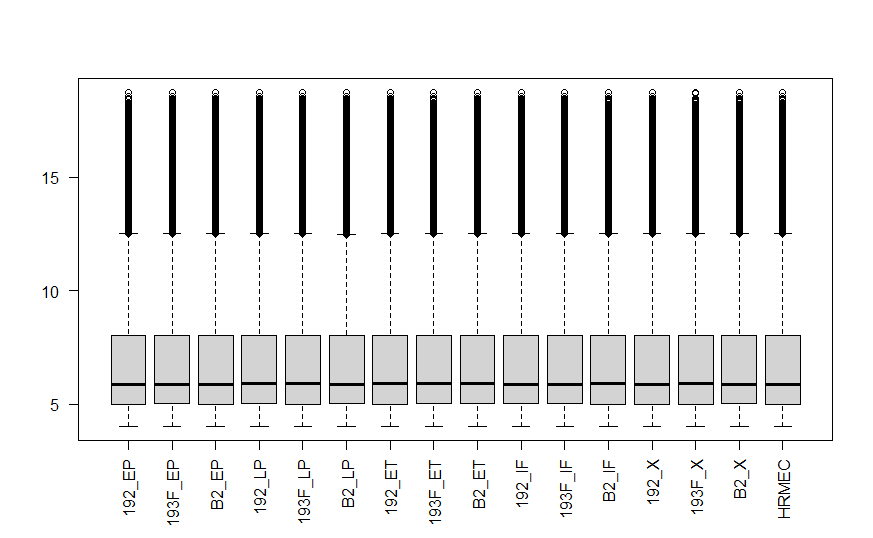
Could you please explain this phenomenon? Thanks.