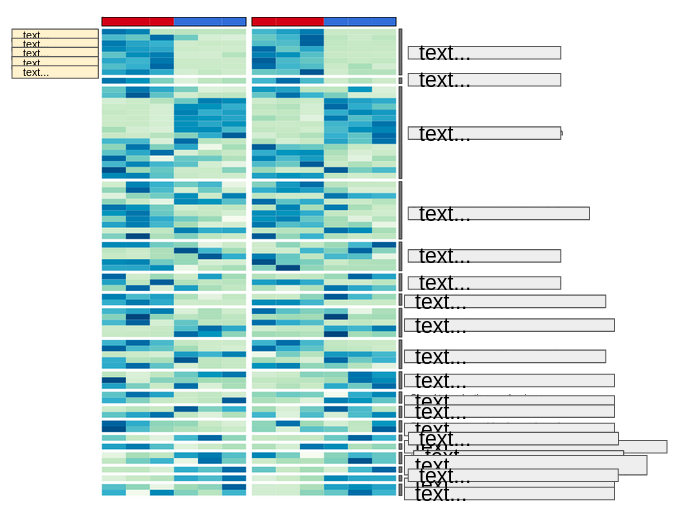
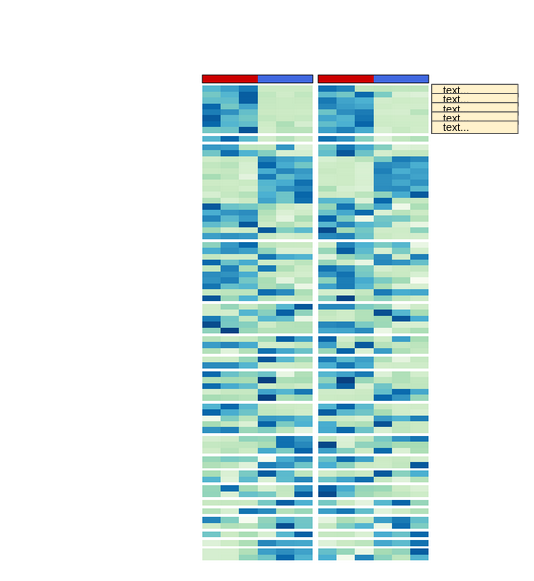
I am drawing two heatmaps, one next to the other, aligned by row names. To achieve this I am using the plus sign (+) to concatenate them. The function works properly and both heatmaps are drown next to each other and aligned.
#-------------------------------------------------------------------------------
# Load Heatmap Packages and complements
#-------------------------------------------------------------------------------
library("ComplexHeatmap")
library("RColorBrewer")
library("circlize")
draw(
myLogHeatMap + myStatHeatMap,
padding = unit(c(10, 10, 10, 10), "mm")
)
myLogHeatMap has a left_annotation and myStatHeatMap has a right_annotation:
myStatHeatMap <- Heatmap(
### 1) Heatmap Matrix
matrix = myScaledStatMatrix,
### 2) Color of the Heatmap body
col = hmcol,
### 3) Annotations
top_annotation = stat_column_labels,
right_annotation = COG_annotations,
...
myLogHeatMap <- Heatmap(
### 1) Heatmap Matrix
matrix = myScaledLogMatrix,
### 2) Color of the Heatmap body
col = hmcol,
### 3) Annotations
top_annotation = log_column_labels,
left_annotation = geneAnno,
...
But I need them to be inverted, the right_annotation should be the left_annotation:
myStatHeatMap <- Heatmap(
### 1) Heatmap Matrix
matrix = myScaledStatMatrix,
### 2) Color of the Heatmap body
col = hmcol,
### 3) Annotations
top_annotation = stat_column_labels,
right_annotation = geneAnno,
...
myLogHeatMap <- Heatmap(
### 1) Heatmap Matrix
matrix = myScaledLogMatrix,
### 2) Color of the Heatmap body
col = hmcol,
### 3) Annotations
top_annotation = log_column_labels,
left_annotation = COG_annotations,
...
But I obtain a blank space instead of the COG annotations text labels:
and the following error at the console:
Error in grid.Call.graphics(C_downviewport, name$name, strict) :
Viewport 'annotation_COG_1' was not found
In addition: Warning message:
These are the annotation constructors I used, they worked ok until I swap the right and left_annotation:
#-------------------------------------------------------------------------------
# row names annotations
#-------------------------------------------------------------------------------
geneAnno <- rowAnnotation(rows = anno_text(
rownames(myScaledStatMatrix),
rot = 0,
gp = gpar(
fontsize = fontsizes,
fontface = fontfaces,
col = fontcolors)
))
#-------------------------------------------------------------------------------
# COG annotations
#-------------------------------------------------------------------------------
COG_annotations = rowAnnotation(
COG = anno_empty(border = FALSE,
width = max_text_width(unlist(COG_list)) + unit(4, "mm") ))
and this is what I used to draw both heatmaps and then the command I used to decorate the left_annotation:
#///////////////////////////////////////////////////////////////////////////////
# Draw the two Heatmaps, their Legends, and Annotations
#///////////////////////////////////////////////////////////////////////////////
draw(
myStatHeatMap + myLogHeatMap,
padding = unit(c(10, 10, 10, 10), "mm")
)
#-------------------------------------------------------------------------------
# Add the COG Labels decorations
#-------------------------------------------------------------------------------
for (i in 1:19) {
decorate_annotation("COG", slice = i , {
grid.rect(
x = "0",
width = unit(1, "mm"),
gp = gpar(fill = "gray44", col = "gray20"),
just = "left"
)
grid.text(
paste(COG_list[[i]], collapse = "\n"),
x = unit(4, "mm"),
gp = gpar(fontsize = 8, fontface = "plain"),
just = "left"
)
})
}
As I mentioned above, this works fine until I swap the annotations side,
Do you know what may be caussing this issue?
Also, here is my sessioninfo() output:
session_info() ─ Session info ──────────────────────────────────────────────────────────────────────────── setting value version R version 4.1.0 (2021-05-18) os Ubuntu 20.04.2 LTS system x86_64, linux-gnu ui RStudio language en_IL:en collate en_IL.UTF-8
ctype en_IL.UTF-8 tz Asia/Jerusalem
date 2021-07-12─ Packages ──────────────────────────────────────────────────────────────────────────────── package * version date lib BiocGenerics
0.38.0 2021-05-19 1 cachem 1.0.5 2021-05-15 1 Cairo 1.5-12.2 2020-07-07 1 callr 3.7.0 2021-04-20 1 circlize
- 0.4.13 2021-06-09 1 cli 3.0.0 2021-06-30 1 clisymbols 1.2.0 2017-05-21 1 clue 0.3-59 2021-04-16 1 cluster
2.1.2 2021-04-17 [4] codetools 0.2-18 2020-11-04 [4] colorspace 2.0-2 2021-06-24 1 ComplexHeatmap 2.9.1 2021-07-07 1 crayon
1.4.1 2021-02-08 1 curl 4.3.2 2021-06-23 1 desc 1.3.0 2021-03-05 1 devtools 2.4.2 2021-06-07 1 digest
0.6.27 2020-10-24 1 doParallel 1.0.16 2020-10-16 1 ellipsis 0.3.2 2021-04-29 1 evaluate 0.14 2019-05-28 1 fastmap
1.1.0 2021-01-25 1 foreach 1.5.1 2020-10-15 1 fs 1.5.0 2020-07-31 1 GetoptLong 1.0.5 2020-12-15 1 GlobalOptions
0.1.2 2020-06-10 1 glue 1.4.2 2020-08-27 1 htmltools 0.5.1.1 2021-01-22 1 httpuv 1.6.1 2021-05-07 1 httr
1.4.2 2020-07-20 1 InteractiveComplexHeatmap 1.0.0 2021-05-19 1 IRanges 2.26.0 2021-05-19 1 iterators 1.0.13 2020-10-15 1 jsonlite
1.7.2 2020-12-09 1 kableExtra 1.3.4 2021-02-20 1 knitr 1.33 2021-04-24 1 later 1.2.0 2021-04-23 1 lifecycle
1.0.0 2021-02-15 1 magrittr 2.0.1 2020-11-17 1 matrixStats 0.59.0 2021-06-01 1 memoise 2.0.0 2021-01-26 1 mime
0.11 2021-06-23 1 munsell 0.5.0 2018-06-12 1 pkgbuild 1.2.0 2020-12-15 1 pkgload 1.2.1 2021-04-06 1 png
0.1-7 2013-12-03 1 prettyunits 1.1.1 2020-01-24 1 processx 3.5.2 2021-04-30 1 promises 1.2.0.1 2021-02-11 1 ps
1.6.0 2021-02-28 1 purrr 0.3.4 2020-04-17 1 R6 2.5.0 2020-10-28 1 RColorBrewer 1.1-2 2014-12-07 1 Rcpp
1.0.7 2021-07-07 1 remotes 2.4.0 2021-06-02 1 rjson 0.2.20 2018-06-08 1 rlang 0.4.11 2021-04-30 1 rmarkdown
2.9 2021-06-15 1 rprojroot 2.0.2 2020-11-15 1 rstudioapi 0.13 2020-11-12 1 rvest 1.0.0 2021-03-09 1 S4Vectors
0.30.0 2021-05-19 1 scales 1.1.1 2020-05-11 1 sessioninfo 1.1.1 2018-11-05 1 shape 1.4.6 2021-05-19 1 shiny
1.6.0 2021-01-25 1 stringi 1.6.2 2021-05-17 1 stringr 1.4.0 2019-02-10 1 svglite 2.0.0 2021-02-20 1 systemfonts
1.0.2 2021-05-11 1 testthat 3.0.4 2021-07-01 1 usethis * 2.0.1 2021-02-10 1 viridisLite 0.4.0 2021-04-13 1 webshot
0.5.2 2019-11-22 1 withr 2.4.2 2021-04-18 1 xfun 0.24 2021-06-15 1 xml2 1.3.2 2020-04-23 1 xtable
1.8-4 2019-04-21 1 source Bioconductor CRAN (R 4.1.0)
CRAN (R 4.1.0) CRAN (R 4.1.0)
CRAN (R 4.0.5) CRAN (R 4.0.3)
CRAN (R 4.1.0) Github (jokergoo/ComplexHeatmap@1bd27f8) CRAN (R 4.1.0)
CRAN (R 4.1.0) CRAN (R 4.1.0)
Bioconductor Bioconductor
CRAN (R 4.1.0) CRAN (R 4.1.0)
...1 /home/lgabriel/R/x86_64-pc-linux-gnu-library/4.1 2 /usr/local/lib/R/site-library [3] /usr/lib/R/site-library [4] /usr/lib/R/library