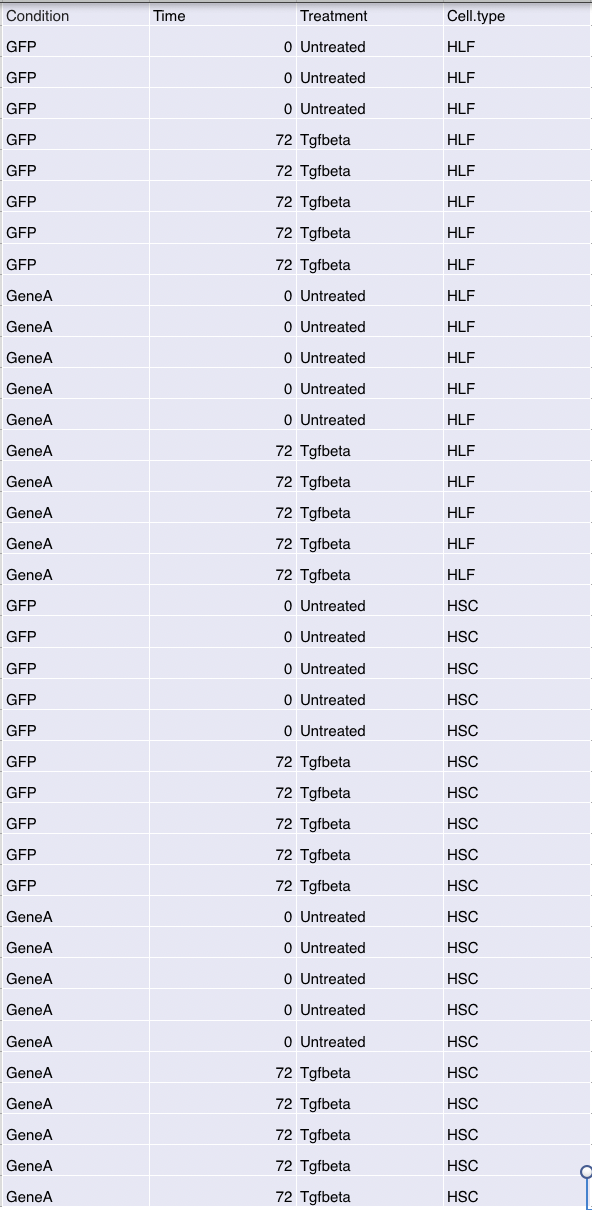
Hi,
Below is the design for the experiment that I am analyzing and essentially GFP is the control. Essentially I have 6 contrasts 1) 0 hour GFP vs GeneA in HLF 2) 72 hour GFP vs GeneA in HLF 3) 0 hour GFP vs GeneA in HSC 4) 72 hour GFP vs GeneA in HSC 5) 0 hour GFP vs GeneA in HSC vs HLF 6) 72 hour GFP vs GeneA in HSC vs HLF
I am unsure how to go about doing the design and contrasts for 5) and 6). Essentially I would like to identify genes showing different patterns between GFP and GeneA in HSC vs HLFs in limma (i.e. increasing in HSCs and decreasing or not changing in HLFs and vice versa). Is this type of contrast even possible in limma if so how would one do this type of contrast and design?
Kind Regards,
Sam