Entering edit mode
Hello Jianhong,
I was using trackViewer to generate plotting for ChIPSeq data. In the end result, the font on the labeling for each track seem too big, e.g. 'genes', 'H1', 'H2'. I can change these after exporting to PDF and edit by AI. But, is there a way to change font size in R code?
Thanks!
optSty <- optimizeStyle(trackList(HA1, HA2, genes), theme="safe")
trackList <- optSty$tracks
viewerStyle <- optSty$style
setTrackViewerStyleParam(viewerStyle, "margin", c(.05, .2, .05, .05)) #bottom, left, top and right margin.
setTrackXscaleParam(trackList[[2]], "draw", TRUE)
setTrackXscaleParam(trackList[[2]], "gp", list(cex=0.8))
# setTrackXscaleParam(trackList[[2]], attr="position",
# value=list(x=69035856, y=2, label=1000))
setTrackViewerStyleParam(viewerStyle, "xaxis", FALSE) # disable x ticks
setTrackStyleParam(trackList[[1]], "ylim", c(0, 0.5))
setTrackStyleParam(trackList[[2]], "ylim", c(0, 0.5))
vp <- viewTracks(trackList,
gr=gr, viewerStyle=viewerStyle,
autoOptimizeStyle=TRUE)
addGuideLine(c(69035856, 69037005), vp=vp) #peak1
addArrowMark(list(x=(69035856 + 69037005)/2,
y=2), # 2 means track 2 from the bottom.
label="peak",
col="darkgreen",
vp=vp)

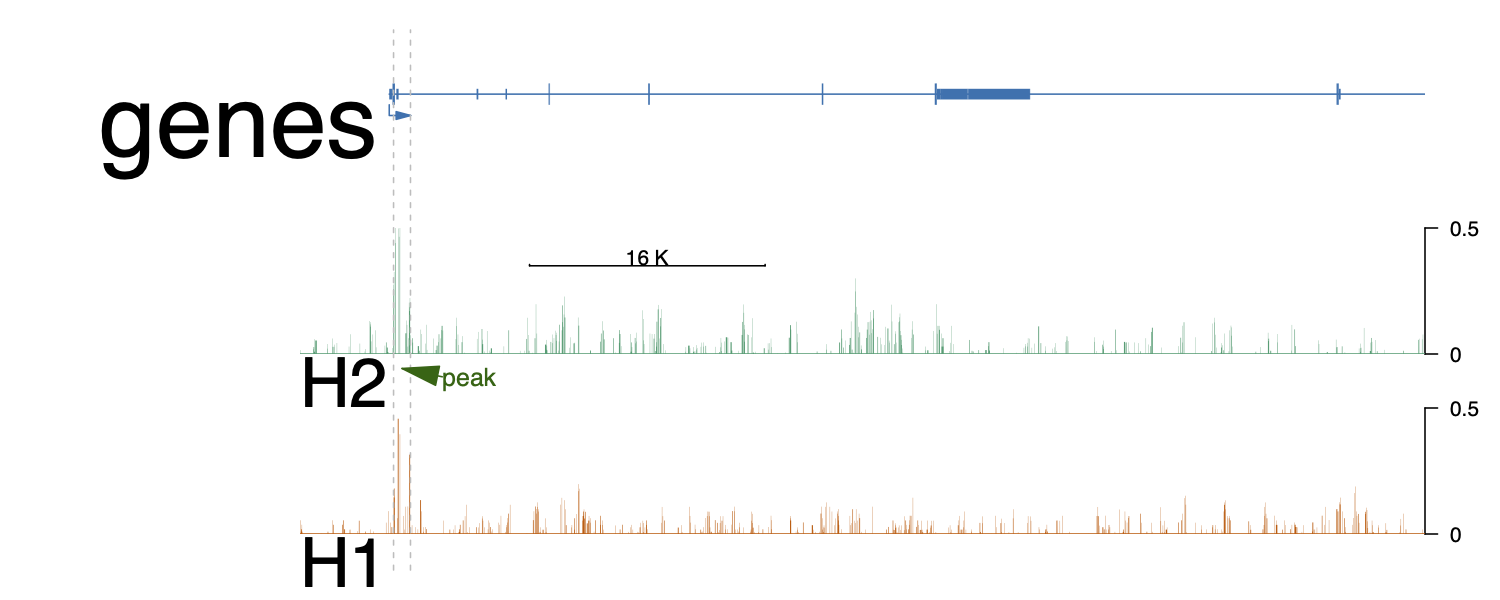

Thanks! This adjustment works very well