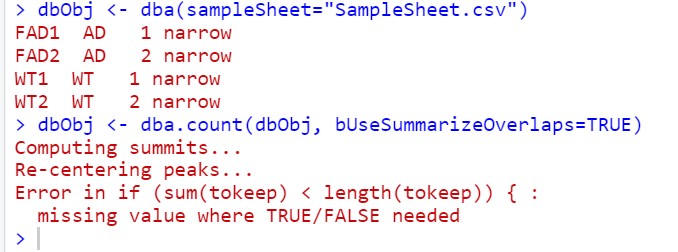
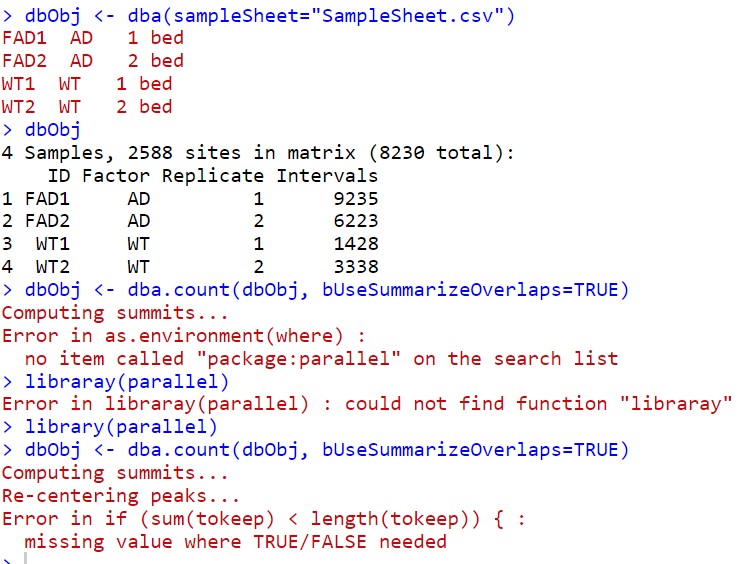
When I used dba.cout with DiffBind, the following error occurred.
Code should be placed in three backticks as shown below
library(DiffBind)
library(parallel)
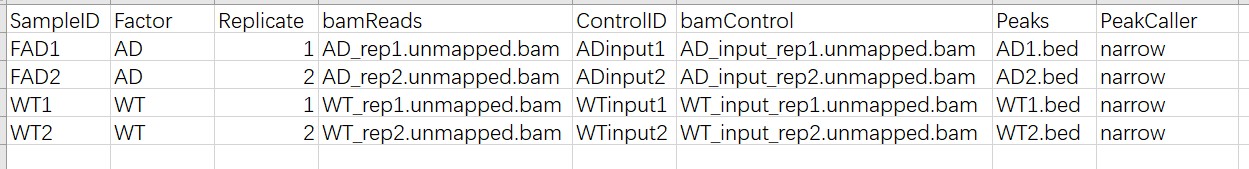
dbObj <- dba(sampleSheet="SampleSheet.csv")
dbObj <- dba.count(dbObj, bUseSummarizeOverlaps=TRUE)
The following error occurred
Error in if (sum(tokeep) < length(tokeep)) { :
missing value where TRUE/FALSE needed
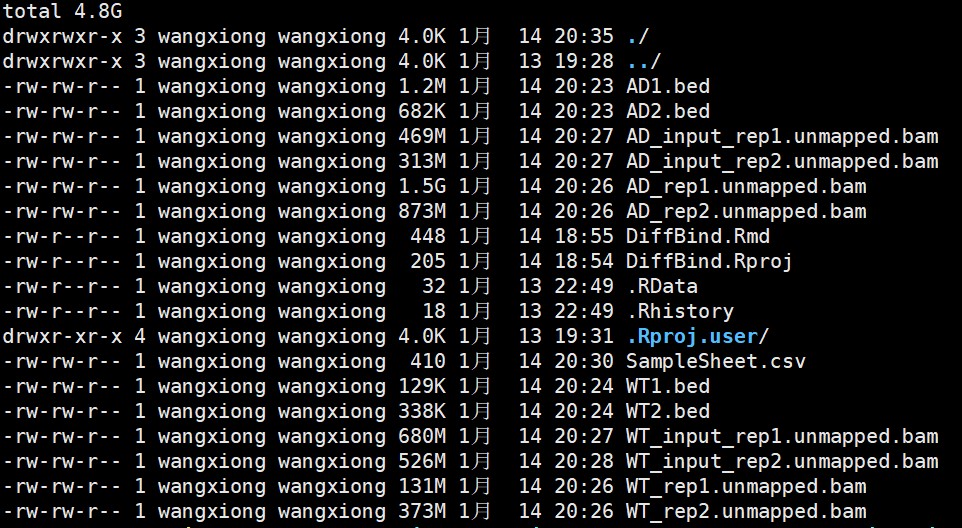
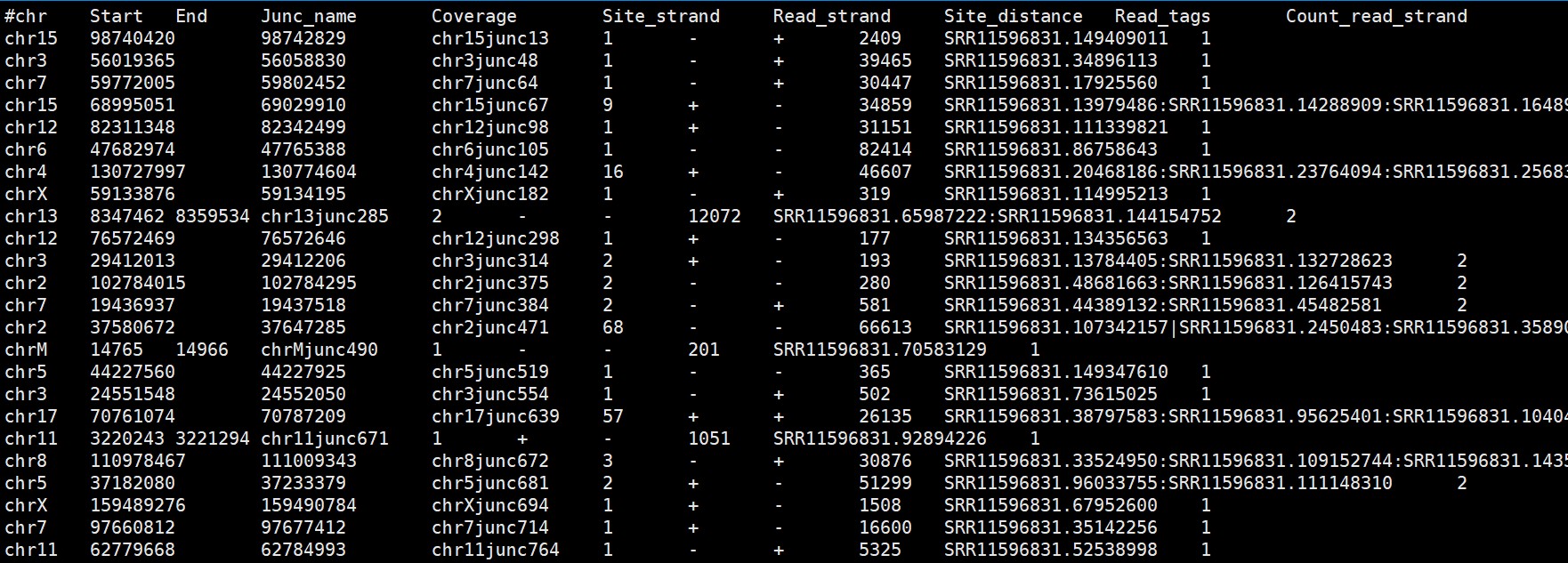
The bam files were obtained after bowtie2 alignment, and the bed files were obtained using AutoCirc. The last line of these bed files was not truancated.
Here are some related figures.
Here is the head of bed file


Can anyone help?