Need help, I'm having issue since prostar v1.26.4 with data that were working previously.
The problem seems to be with converting the maxquant data (Same file has been used recently with prostar 1.26.2 and 1.26.3 with success)
I get this warning in R after the convert step:
"Warning in xtfrm.data.frame(x) : cannot xtfrm data frames "
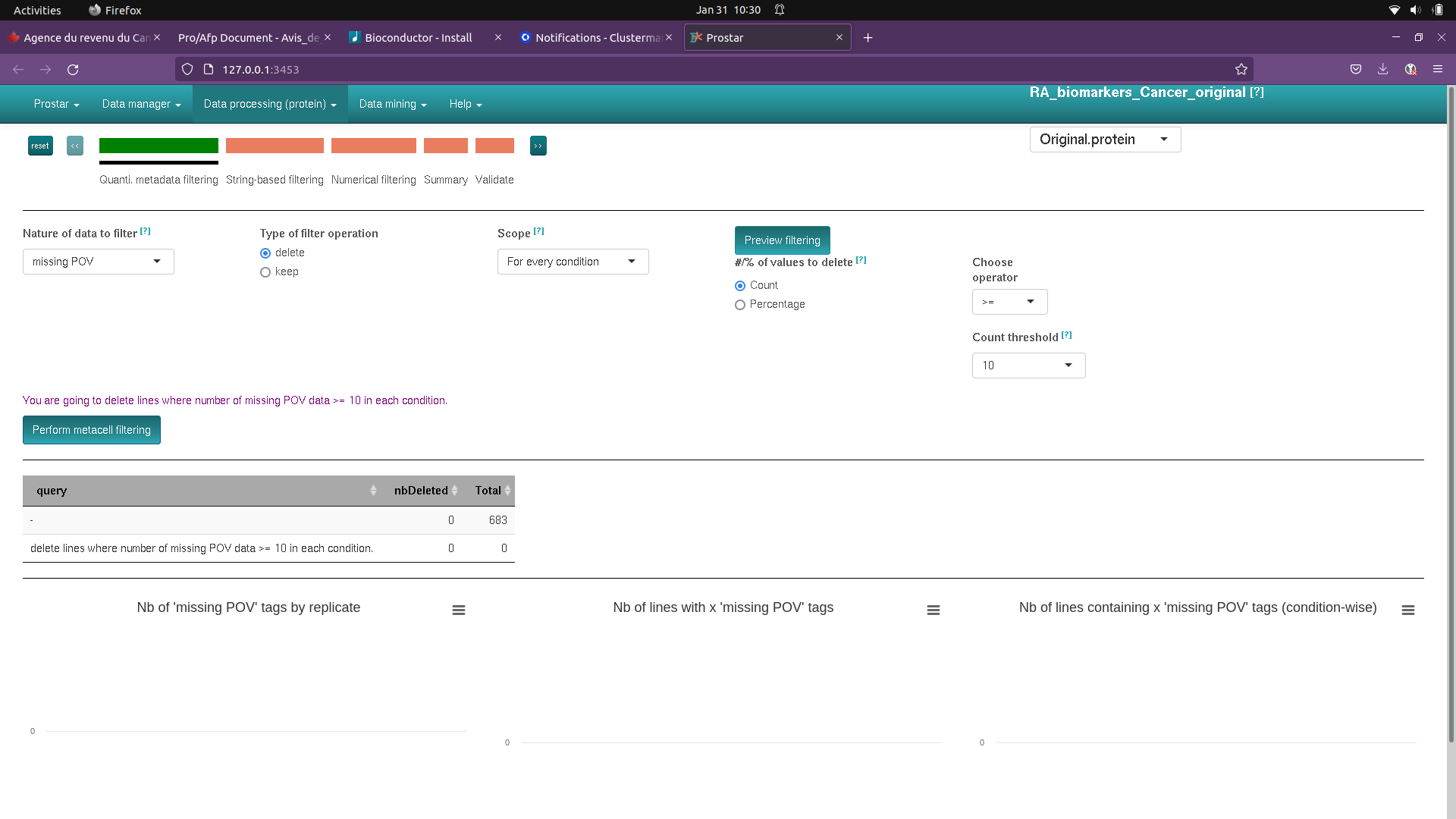
If I go through filtering it looks like all my data lines are lost (see screenshot)
I get the following warnings in R
"Warning in metacellHisto_HC(obj = obj(), pattern = pattern(), pal = pal()) :
'pattern' is empty.
Warning in metacellPerLinesHisto_HC(obj = obj(), pattern = pattern(), indLegend = c(2:length(colnames(Biobase::pData(obj()))))) :
'pattern' is empty.
Warning in metacellPerLinesHistoPerCondition_HC(obj = obj(), pattern = pattern(), :
'pattern' is empty.
Warning in metacellHisto_HC(obj = obj(), pattern = pattern(), pal = pal()) :
'pattern' is empty.
Warning in metacellPerLinesHisto_HC(obj = obj(), pattern = pattern(), indLegend = c(2:length(colnames(Biobase::pData(obj()))))) :
'pattern' is empty.
Warning in metacellPerLinesHistoPerCondition_HC(obj = obj(), pattern = pattern(), :
'pattern' is empty."
I've encountered the same error running the zero-install v1.26.4 in a windows 10 VM.
Could this be an issue in update v1.26.4 ?
Thanks
sessionInfo( )
R version 4.1.2 (2021-11-01) Platform: x86_64-pc-linux-gnu (64-bit) Running under: Ubuntu 21.10
Matrix products: default BLAS: /usr/local/lib/R/lib/libRblas.so LAPACK: /usr/local/lib/R/lib/libRlapack.so
locale:
1 LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8
[6] LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages: 1 stats graphics grDevices utils datasets methods base
other attached packages: 1 Prostar_1.26.4 BiocManager_1.30.16
loaded via a namespace (and not attached):
1 compiler_4.1.2 tools_4.1.2
```