I'm using ArchR to analyze a H.sapiens PBMC scATAC dataset I have and decided to use Ensembl's GRCh38, Release 103 genome as my reference.
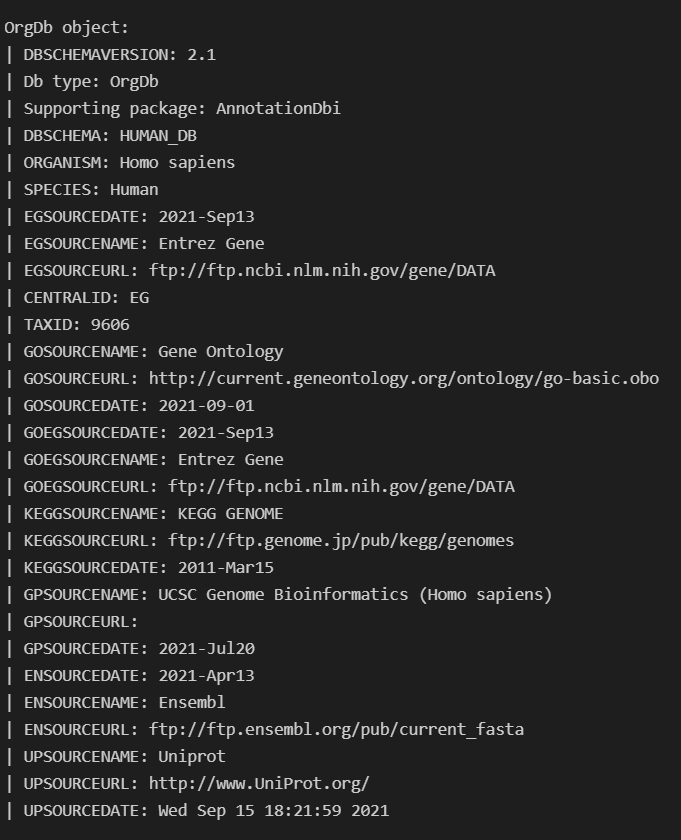
In order to do this I needed to use ArchR's createGenomeAnnotation & createGeneAnnotation and to define the genome. For createGeneAnnotation an OrgDb object was need which I used AnnotationHub to access.
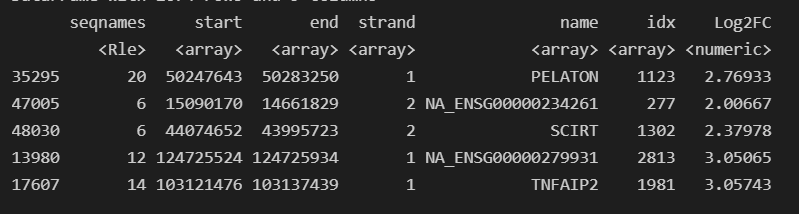
Once I loaded in this OrgDb I realized there were GENEIDs (EntrezIDs) present in my GTF for GRCh38, Release 103 that were missing from the OrgDb. I thought this meant it wasn't up to date, but realized only one was returned when I queried annotation hub for it like this query(hub, c("Homo sapiens","OrgDb")).
Is there somewhere I can get a more up to date version of the H.sapiens OrgDb? I was under the impression it was regularly updated so had trouble believing that it was missing so many IDs
Its resulting in detected marker genes being labeled as NA_<GeneIDPresentInGTF>
Any and all guidance would be greatly appreciated


Cross-posted to Biostars https://www.biostars.org/p/9513406/