Entering edit mode
Hi everyone,
Generating a pheatmap for my analysis. Everything is going to plan except for the colouring of the map itself - only some samples are being coloured whilst the rest are blank.
Here is my script:
sampledist <- dist(t(assay(vsd)), method = "euclidean")
sampledist_matrix <- as.matrix(sampledist)
colors <- colorRampPalette(rev(brewer.pal(9, "YlGnBu")))(255)
Specify colours for experimental design annotations:
variety_clr <- brewer.pal(3, "Set3")[c(1,3)]
names(variety_clr) <- unique(coldata$Variety)
time_clr <- brewer.pal(4, "Set1")
names(time_clr) <- unique(coldata$Time)
annotation_colours <- list(Tissue = c(Leaf = "forestgreen", Root = "red"),
Variety = variety_clr, Time = time_clr)
# plot
phm_ed <- pheatmap(sampledist_matrix,
clustering_distance_rows=sampledist,
clustering_distance_cols=sampledist,
clustering_method = "average",
treeheight_row = 80,
treeheight_col = 80,
annotation_col = coldata[2:4,],
annotation_row = coldata[2:4,],
annotation_colors = annotation_colours,
show_rownames = TRUE,
show_colnames = FALSE,
cellwidth = 15,
cellheight = 15,
cutree_rows = 2,
cutree_cols = 2,
col=colors,
fontsize = 12,
border_color = "black")
If anyone could shed any light as to why this might be I would appreciate the help.
Thanks in advance

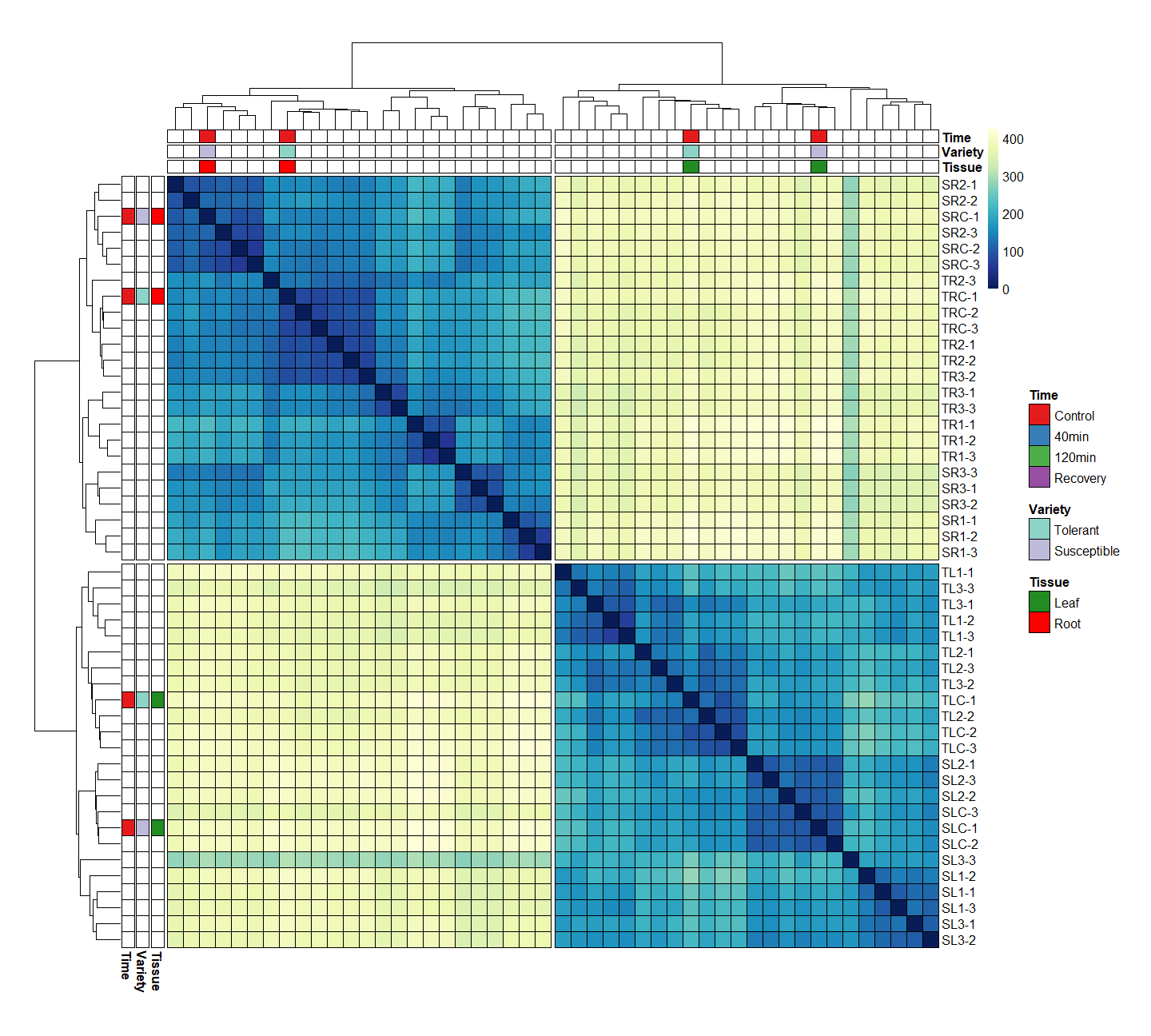

Does the ":" between the 2 and the 4 not select for columns 2, 3, and 4? That was my impression but I am new to this. What would the alternative be if not this? Thanks!
It does select for 2, 3, and 4. But [2:4,] is rows 2-4. If you want columns, it's [,2:4]. Note which side of the comma the range is on.
Ah great I see - I've managed to solve this issue now. Thanks again.