Entering edit mode
Hey everyone,
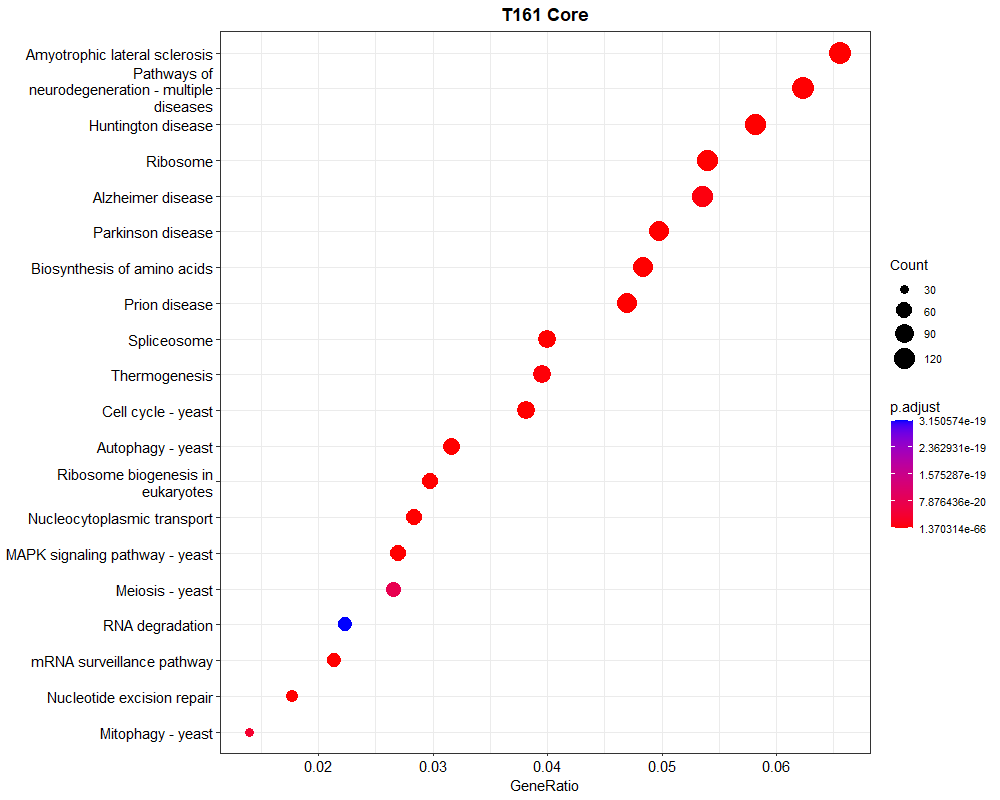
In order to do gene enrichment analysis on non-model organism (fungi), I obtained the KEGG ids using KOfam analysis. Using clusterProfiler enrichKEGG function I did the enrichment analysis. But the problem now is the enriched pathways I am getting is human diseases like Parkinsons disease, Huntington diseases etc. or human related pathways. Is there a way to filter these out or I am doing something wrong. My codes are given below:
file <- read.delim("T161_core.txt", header = FALSE, sep="\t")
colnames(file) <- c("ProteinID", "KO")
file1<- (file$KO)
kegg <- enrichKEGG( file1, organism = "ko", keyType = "kegg", pvalueCutoff = 0.05,
pAdjustMethod = "BH", qvalueCutoff = 0.05)
p<- dotplot(kegg, showCategory=20) + ggtitle("T161 Core ")
p + theme(
plot.title = element_text(color="black", size=14, face="bold", hjust=0.5))
sessionInfo( )


If anyone still wondering about this:
You can filter your 'kegg' object based on a list of target pathways.
Look at these fields:
kegg@result[["Description"]], which has pathway namesand
kegg@result[["ID"]], which has respective pathway IDscan you tell me the formate of output file from KOfam please? I don't understand the formate of input file for ClusterProfiler.
Hello, I'm also analyzing a non-model organism. Could you tell me which input files you used? Also, why did you set `organism = 'ko' ?