Dear rnaseqDTU & DRIMseq community,
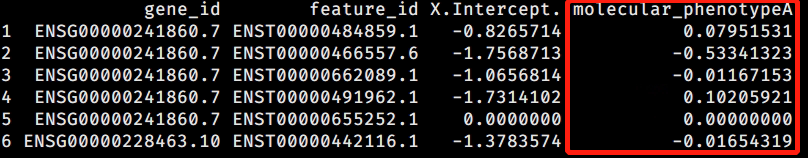
I am performing Swimming downstream , and I want to confirm those transcripts overused in condition A compared with condition B. So I went looking for some metrics similar to logFC in DESeq2 and I found coefficient() function on dmDStest could give me a 'regression coefficient' for each transcript
I'm not statistician and I hope I understand it correctly, that:
- coefficient(dmDStest) gives trasncript level coefficient using Dirichlet-multinominal (DM) model,
- coefficient(dmDStest, level = "feature") gives trasncript level coefficient using Beta-Binominal (BB) model
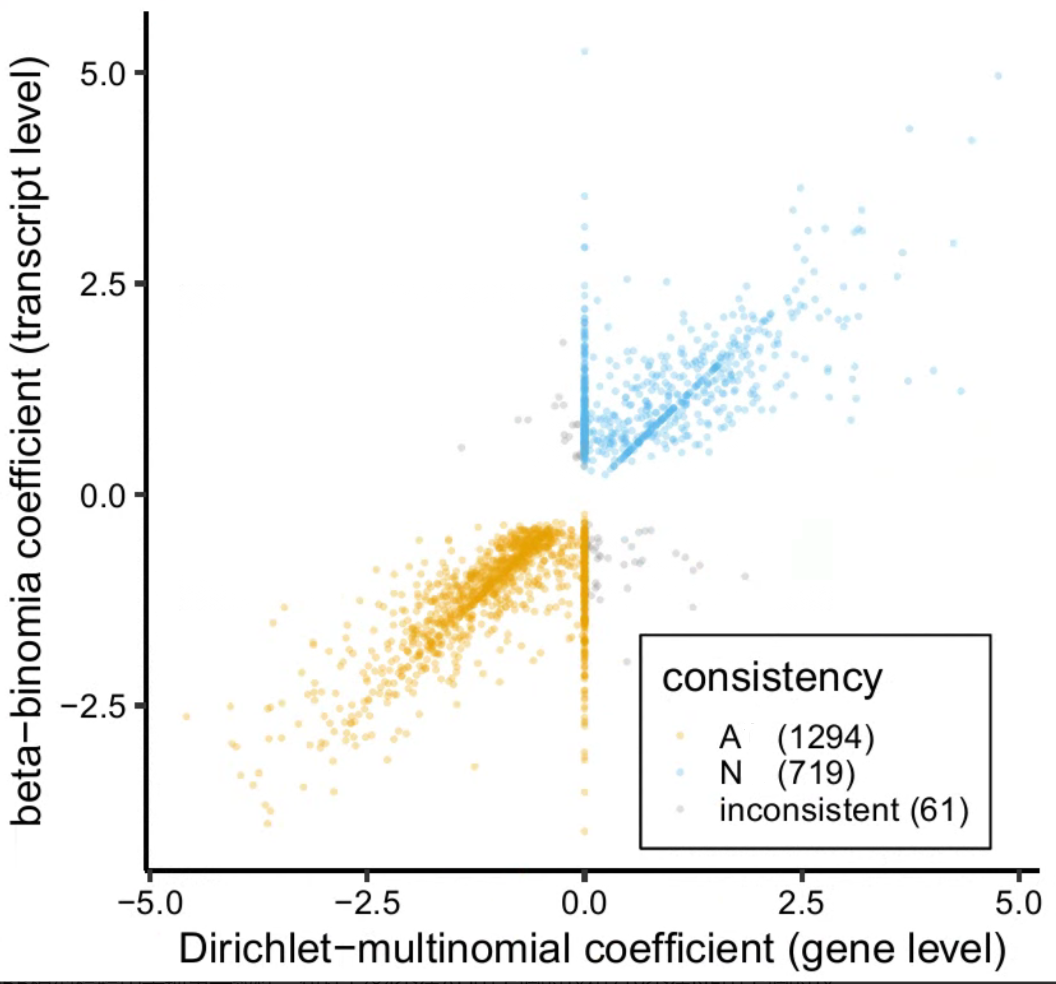
then I extracted all transcripts with a padj < 0.05 (follow this post) and plot DM coef and BB coef at x- and y-axis. I found:
- many transcripts showed DM coef that equal 0
- some transcripts showed DM coef and BB coef with incosistent sign.
I would like to ask:
- if it using the sign of the coef to define the direction of DTU in different condition resonable (just like log2FC in DESeq2 analysis)
- If resonable, which coef should I use to determine the direction of DTU in different condition?
- How to deal with those transcripts with inconsistent coef?
- could I say that the higher the coeffcient, the higher the overusage?
Best regards,
Wang


Thank you so much. Your kind explanation also helps me understand why some transcript have identical coefficient in both models!