Hi All:
I would like to ask a question about interaction term in DESeq2. I have the following design:
NF2 <- factor(c("Ctrl", "Ctrl", "Ctrl", "KnockDown", "KnockDown", "KnockDown", "KnockDown", "KnockDown", "KnockDown"))
PRG <- factor(c("Untreated", "Untreated", "Untreated", "Untreated", "Untreated", "Untreated", "Treated", "Treated", "Treated"))
prg_metadata_G2G3G5 <- data.frame(row.names = colnames(prg_countdata_G2G3G5), NF2, PRG)
dds_G2G3G5 <- DESeqDataSetFromMatrix(countData = prg_countdata_G2G3G5,
colData = prg_metadata_G2G3G5,
design = ~ NF2 + PRG + NF2:PRG)
I have three samples (G2, G3, G5), and each sample has three replicates.
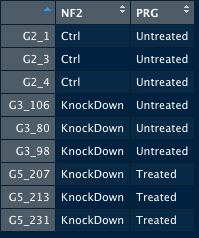
I have two condition, control (Ctrl) and Knockdown, and two treatments (drugs), Untreated and treated. G2 is a control sample, untreated by drug. G3 is a knockdown sample untreated by drug. G5 is a knockdown sample treated by drug.
Thus, colData looks like this:
I am interested in differences in response between treatments. Thus, I used an interaction term as above. However, it gives an error: the model matrix is not full rank, so the model cannot be fit as specified. One or more variables or interaction terms in the design formula are linear combinations of the others and must be removed.
What's wrong with my design formula? How can I fix it?
I'd appreciate any help to the design formula to improve my design to achieve the output.
Thanks.