Hey!
Thank you for a great resource! I am learning how to use DESEQ2 on time-series and I would like to get advice if my analysis is statistically "correct".
I have a single time-series i.e. the first time point is the control (t=-1) and the treatment starts from the second timepoint onwards (t=0). I have total of 8 timepoints of treatment (t=0, 1, 2, 3, 4, 5, 6, 7).
I want to compare each treatment timepoint with the control, and each treatment timepoint with the other. I am able to extract out DESeq results using the following code.
ddsMat <- DESeqDataSetFromMatrix(countData = countdata2,
colData = metadata2,
design = ~ Time)
ddsMat <- DESeq(ddsMat)
#output of each timepoint with "-1"
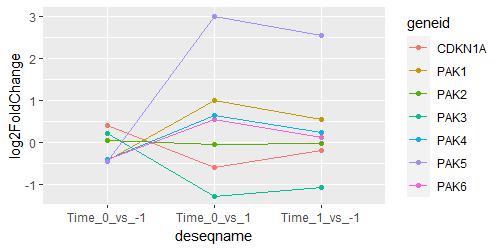
res_1 <- as.data.frame(results(ddsMat, name="Time_0_vs_.1", test="Wald",alpha=0.05,pAdjustMethod = "BH",format="DataFrame"))
res_2 <- as.data.frame(results(ddsMat, name="Time_1_vs_.1", test="Wald",alpha=0.05,pAdjustMethod = "BH",format="DataFrame"))
#-----------------------------------
#condition2-output of each timepoint with the one before-
res2 <- as.data.frame(results(ddsMat, contrast=c("Time","1","0"),test="Wald",alpha=0.05,pAdjustMethod = "BH"))
I am interested in comparing res_1 and res_2, and found search results suggesting using ANOVA to compare these time points. When I examine res2 (called as Time_0_vs_1) for certain genes, I find that their log2FoldChange value lie between res_1 (called as Time_0_vs_-1) and res_2 (called as Time_1_vs_-1). Any suggestions on how to proceed?