Hello everyone,
I would like to know if you could help me about someting in DESeq2.
In the documentation, they talk about technical replicates ad biological replicates. From what I understand:
- biological replicates are for example samples that do not come from the same people/animal/cellular culture.
- technical replicates are for example samples that were sequenced twice on the same or on different machine.
After discussion with other bioinformatician and biologist, it appears to me that sometime we do not have the same consideration of the term "biological replicate". Indeed, If I make some swab from the same animal, does it results in technical replicate (because they come from the same individual) or biological replicates ? (because it's not exactly the same sample that is sequenced twice).
I hope my question is enough clear.
Thanks
Léo


Hi Michael,
Does it means that is it possible to compare for example one treated animal against one untreated animal if I have 2 samples of each animal (and so 2 different library preparation for each animal) ?
For me it does not really reflect the biological variability that DESeq2 requires for the differential testing step. What is your definition of biological replicates?
Thanks,
Léo
That experiment doesn't make sense to me, because you aren't observing the key variation which is how the treatment effects different organisms.
So just because it is not a sequencing technical replicate, doesn't mean it provides sufficient biological replication to make inference on the condition effect.
Does it means that is it possible to compare for example one treated animal against one untreated animal if I have 2 samples of each animal (and so 2 different library pirlo tv apk pure preparation for each animal) ?
For me it does not really reflect the biological variability that DESeq2 requires for the differential testing step. What is your definition of biological replicates?
No, I wouldn't say that you have sufficient information to talk about organism differences if you have two organisms, one from each condition. You've only observed within-organism variance but have no evidence of across-organism differences separate from the condition effect. This seems pretty clear to me though, is there a remaining question?
Hi Michael,
I also have a similar question. I wonder in this case, should I mix all samples from the same animal together as biological replicates or do I need to calculate the average for samples from each animal, and then do the DESeq2 analysis using the average counts for each animal?
Below is my experimental design: 3 genotype, 4 rats per genotype, 3-4 samples isolated from per rat.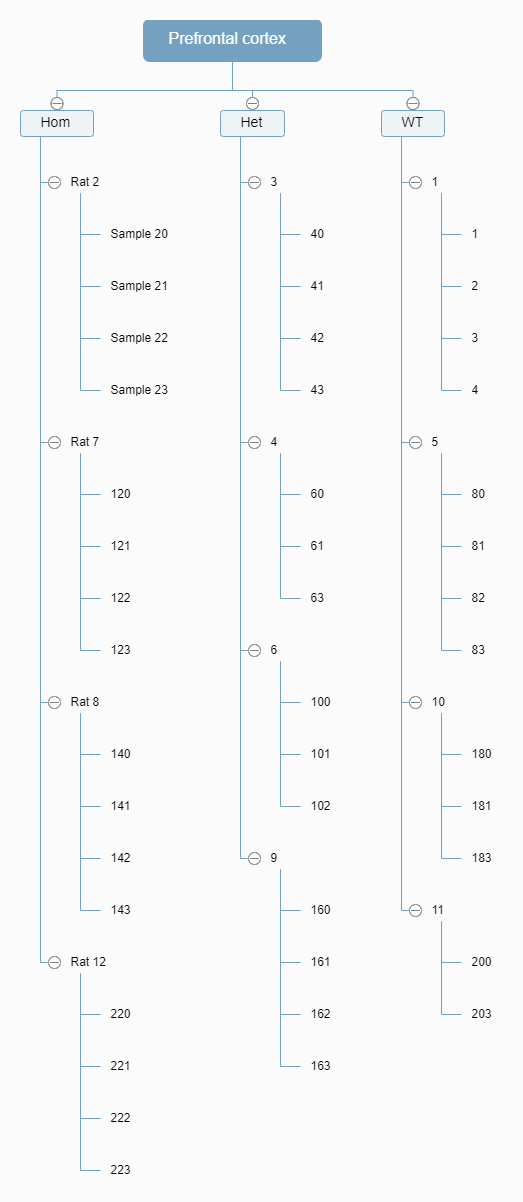
Look forward to your answer. Thank you in advance.
If the multiple replicates are nested within condition, you should use limma with duplicateCorrelation. See related posts on the support site.
Thank you for your reply!
Can I design the formula:
design = ~ animal + genotypeto solve this problem?Not if the animals are nested within genotype.
Many thanks! So there is no function in DESeq2 that can solve this problem?
This has been asked on the support site before. If you look for posts about replicates nested within condition groups you will see me recommend users to use limma with duplicateCorrelation.
https://www.google.com/search?client=firefox-b-1-d&q=site%3Asupport.bioconductor.org+deseq2+nested+condition+duplicatecorrelation
Many thanks!