Entering edit mode
Hi, when I use "GDCprepare" function, I had a such error:
library(TCGAbiolinks)
LUAD_methy <- GDCquery(
project = "TCGA-LUAD",
data.category = "DNA Methylation",
data.type = "Masked Intensities",
platform = "Illumina Human Methylation 450", # Illumina Human Methylation 450
legacy = FALSE)
GDCdownload(LUAD_methy)
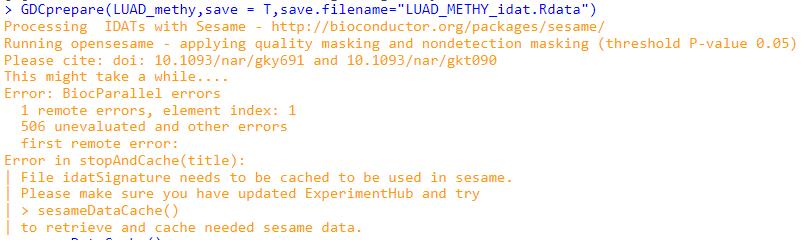
GDCprepare(LUAD_methy,save = T,save.filename="LUAD_METHY_idat.Rdata")
Processing IDATs with Sesame - http://bioconductor.org/packages/sesame/
Running opensesame - applying quality masking and nondetection masking (threshold P-value 0.05)
Please cite: doi: 10.1093/nar/gky691 and 10.1093/nar/gkt090
This might take a while....
Error: BiocParallel errors
1 remote errors, element index: 1
506 unevaluated and other errors
first remote error:
Error in stopAndCache(title):
| File idatSignature needs to be cached to be used in sesame.
| Please make sure you have updated ExperimentHub and try
| > sesameDataCache()
| to retrieve and cache needed sesame data.
When I run sesameDataCache(), it hasn't run for several hours. Is there a good solution? thank you


Do you get something like this? It's waiting for the user to enter "y"
.../Library/Caches/org.R-project.R/R/ExperimentHub does not exist, create directory? (yes/no): y
Hi, I am running into the same issue with the stopAndCache(title):
When I run sesameDataCache() it completes without any errors. However, when I try to read in the sdfs again, I still get the same error. I have updated the ExperimentHub, sesame, and sesameData packages, and tried installing and running the BiocFileCache package as well.
When I run this, it shows that these packages are out of date. Are they relevant to this issue?
Session Info: