Hello everyone,
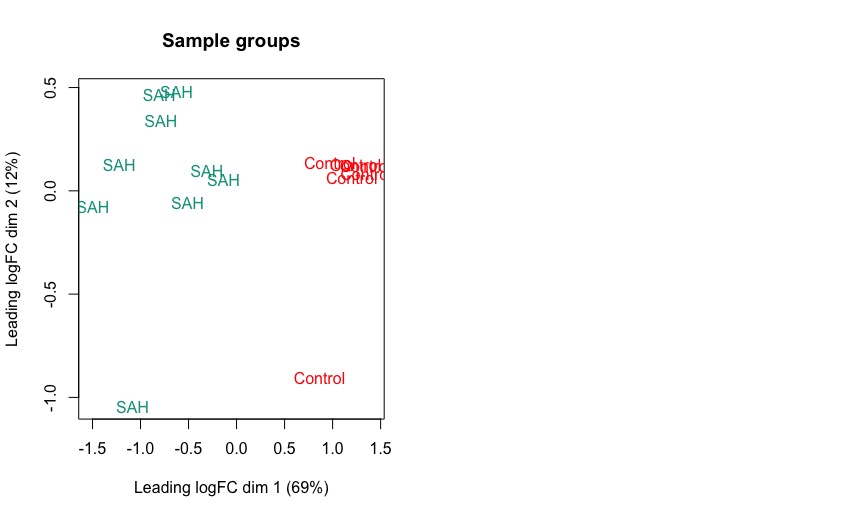
I am struggling with the interpretation of a MDS plot. In my study, I am comparing 84 genes of two different groups (control vs SAH). The exploratory MDS plot groups the SAH condition to the left (negative) on the x axis (leading logFC dim 1) and the control group to the right (positive values). How can I interpret this? I double checked my raw data and it rather looks as if the SAH group tends to have higher log fold changes in many gene compared to the control group (and not smaller logFC as in dim 1 of the graph). The plotMDS function details say that
Distances on the plot can be interpreted as leading log2-fold-change, meaning the typical (root-mean-square) log2-fold-change between the samples for the genes that distinguish those samples.
# Unsupervised clustering of samples
library(wesanderson)
lcpm <- cpm(dge, log=TRUE)
par(mfrow=c(1,2))
col.group <- group
col.group <- as.factor(col.group)
levels(col.group) <- wes_palette(nlevels(col.group), name="Darjeeling1")
col.group <- as.character(col.group)
plotMDS(lcpm, top = 500, labels=group, col=col.group)
title(main="Sample groups")
sessionInfo( )
R version 4.2.1 (2022-06-23)
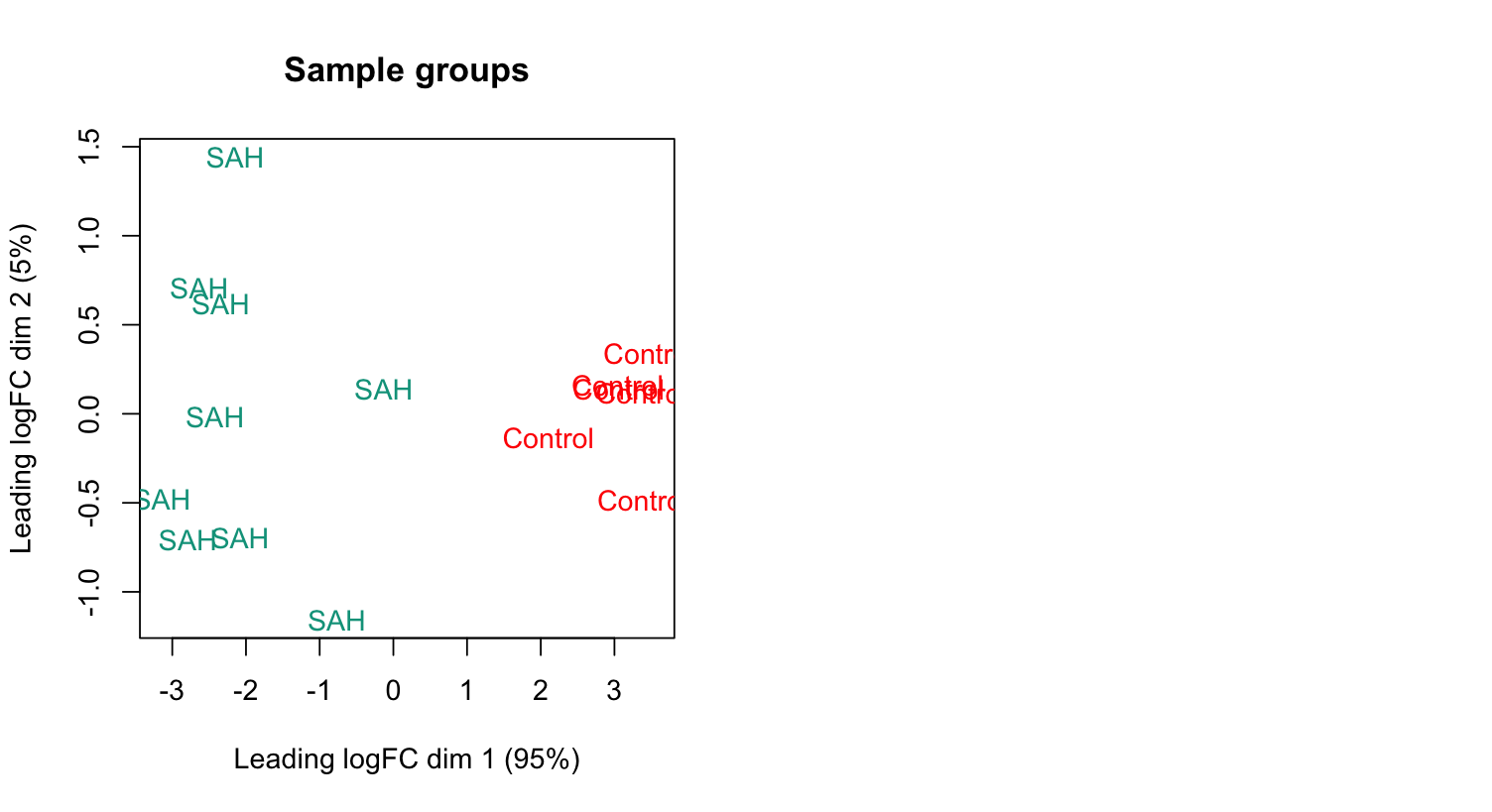
To check this in further detail, I extracted two single differentially expressed genes which are in fact higher expressed in the SAH condition and compared it to the control condition, repeating plotMDS.
sq3=
> par(mfrow=c(1,2))
> col.group <- group
> col.group <- as.factor(col.group)
> levels(col.group) <- wes_palette(nlevels(col.group), name="Darjeeling1")
> col.group <- as.character(col.group)
> plotMDS(sq3, top = 2, labels=group, col=col.group)
> title(main="Sample groups")
Still, logFC dim1 of SAH shows smaller values than the controls.
What am I overlooking (about the code or the calculation behind the plot)?
Thank you so much for your help!



This helped me a lot! Thank you so much.