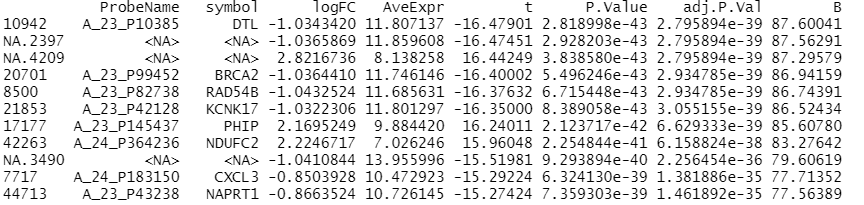
Entering edit mode
Hi, I analyzed an Agilent microarray with limma and wanted to calculate differentially expressed genes for the condition of interest as shown below.
# Differential expression
fit<-lmFit(filt, design)
fit<-eBayes(fit)
results<-decideTests(fit[,2], method='global')
summary(results)
topTable(fit, coef=2, sort.by='p')
The output, however, included some probes labeled as NAs which I can not explain as probes were filtered for those that have a symbol annotation.
Can someone explain what I am doing wrong?
> sessionInfo()
R version 4.1.2 (2021-11-01)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United Kingdom.1252 LC_CTYPE=English_United Kingdom.1252 LC_MONETARY=English_United Kingdom.1252
[4] LC_NUMERIC=C LC_TIME=English_United Kingdom.1252
system code page: 65001
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] org.Hs.eg.db_3.14.0 AnnotationDbi_1.56.2 IRanges_2.28.0 S4Vectors_0.32.3 limma_3.50.0 GEOquery_2.62.2
[7] Biobase_2.54.0 BiocGenerics_0.40.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 XVector_0.34.0 GenomeInfoDb_1.30.0 pillar_1.8.0 compiler_4.1.2
[6] R.methodsS3_1.8.1 R.utils_2.12.2 zlibbioc_1.40.0 bitops_1.0-7 tools_4.1.2
[11] bit_4.0.4 RSQLite_2.2.9 memoise_2.0.1 lifecycle_1.0.1 tibble_3.1.8
[16] png_0.1-7 pkgconfig_2.0.3 rlang_1.0.4 DBI_1.1.1 cli_3.1.0
[21] rstudioapi_0.13 curl_4.3.2 fastmap_1.1.0 GenomeInfoDbData_1.2.7 httr_1.4.2
[26] dplyr_1.0.9 xml2_1.3.3 Biostrings_2.62.0 generics_0.1.3 vctrs_0.4.1
[31] hms_1.1.1 bit64_4.0.5 tidyselect_1.1.2 glue_1.6.2 data.table_1.14.2
[36] R6_2.5.1 fansi_0.5.0 tzdb_0.2.0 readr_2.1.2 purrr_0.3.4
[41] tidyr_1.1.4 blob_1.2.2 magrittr_2.0.1 ellipsis_0.3.2 KEGGREST_1.34.0
[46] assertthat_0.2.1 utf8_1.2.2 RCurl_1.98-1.5 cachem_1.0.6 crayon_1.4.2
[51] R.oo_1.24.0
Thanks in advance!