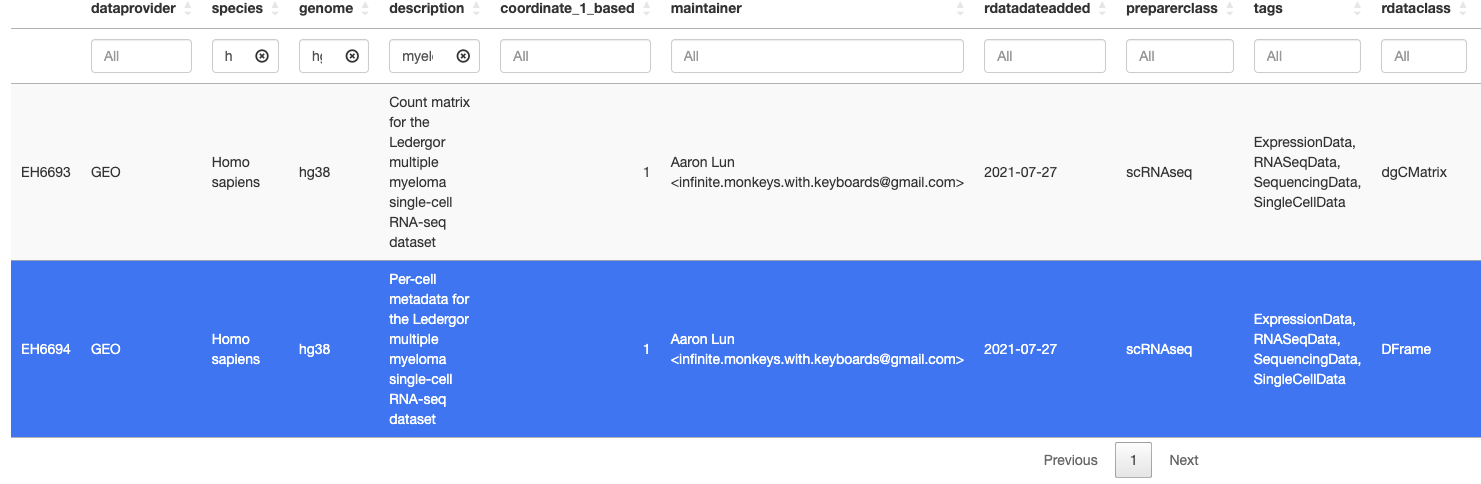
I would like to annotate or label UMAP clusters with cell types in a multiple myeloma single-cell RNA-seq dataset using the scRNAseq R package in conjunction with the SingleR package, if possible. I found a dataset that I would like to use on the scRNAseq dataset collection which is in the LedergorMyelomaData() function which returns a SingleCellExperiment object. I notice that there is no ''cell label'' column within this object? Does that mean I cannot use this data as a reference to annotate my own myeloma dataset's clusters? In terms of cell labels, does it depend on each dataset in the scRNAseq collection if it has the labels there or not? I also searched through the ExperimentHub() repository and found the same dataset which seems to be under the code EH6694 and is shown in the below screenshot.
If, it is not possible to use the Ledergor data for cell type annotation, what dataset would you recommend using instead? I would like to compare to a public reference if possible.
library(scRNAseq)
#
mm_annotations <- LedergorMyelomaData()
#
head(mm_annotations@colData)