Entering edit mode
I want to plor rarefaction cruves from a phyloseq object made from QIIME2 objects:
otu_table = Biomtable from qiime2
Tax_table = taxonomic assign in tsv format
sample_data = metadata in tsv format
I want to rarefy my data, but i need to generate rarefaction curves and i don´t know how to do it. I tried with other packages from Github but I can´t by the moment :(
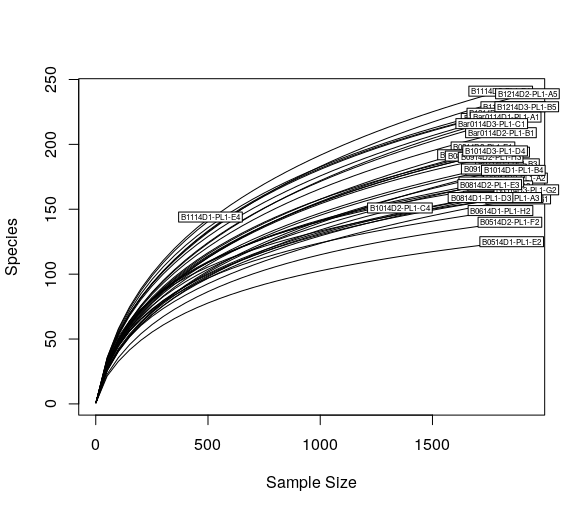
Iwant to plot sth like this:
Thanks


I am not sure what you have tried, but the "vegan" library (package) has the ability to do rarefaction. So make sure to load
library("vegan")and then something likerarecurve(BCI, step = 20, sample = raremax, col = "blue", cex = 0.6). Documentation for rarefy can be found here. In this tutorial we can see they userarecurve()for phyloseq data, there they usedrarecurve(t(otu_table(ps)), step=50, cex=0.5)but usingrarecurve()from vegan should allow you to generate a rarefaction curve from the phyloseqotu_table.