Entering edit mode
Hello,
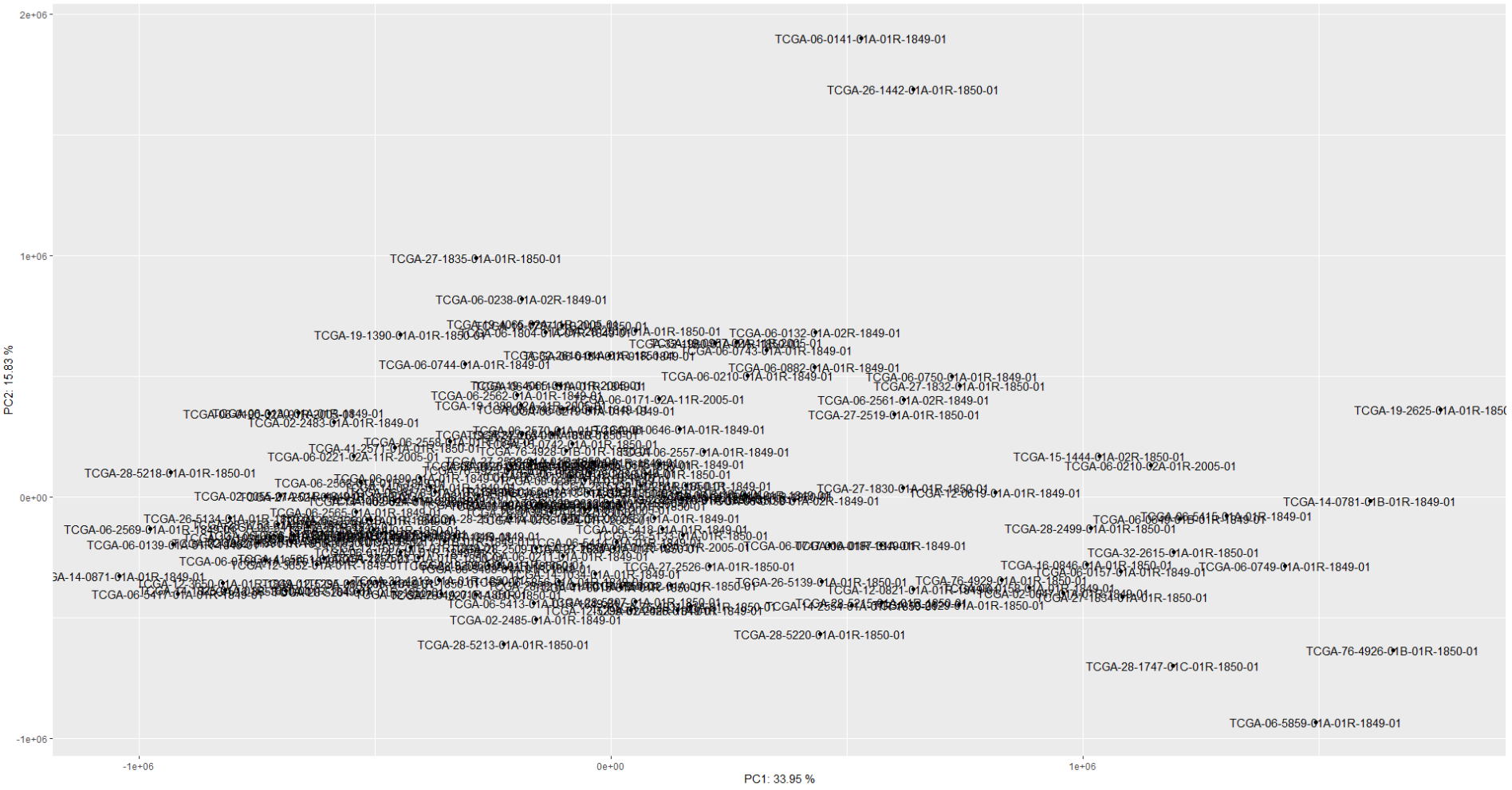
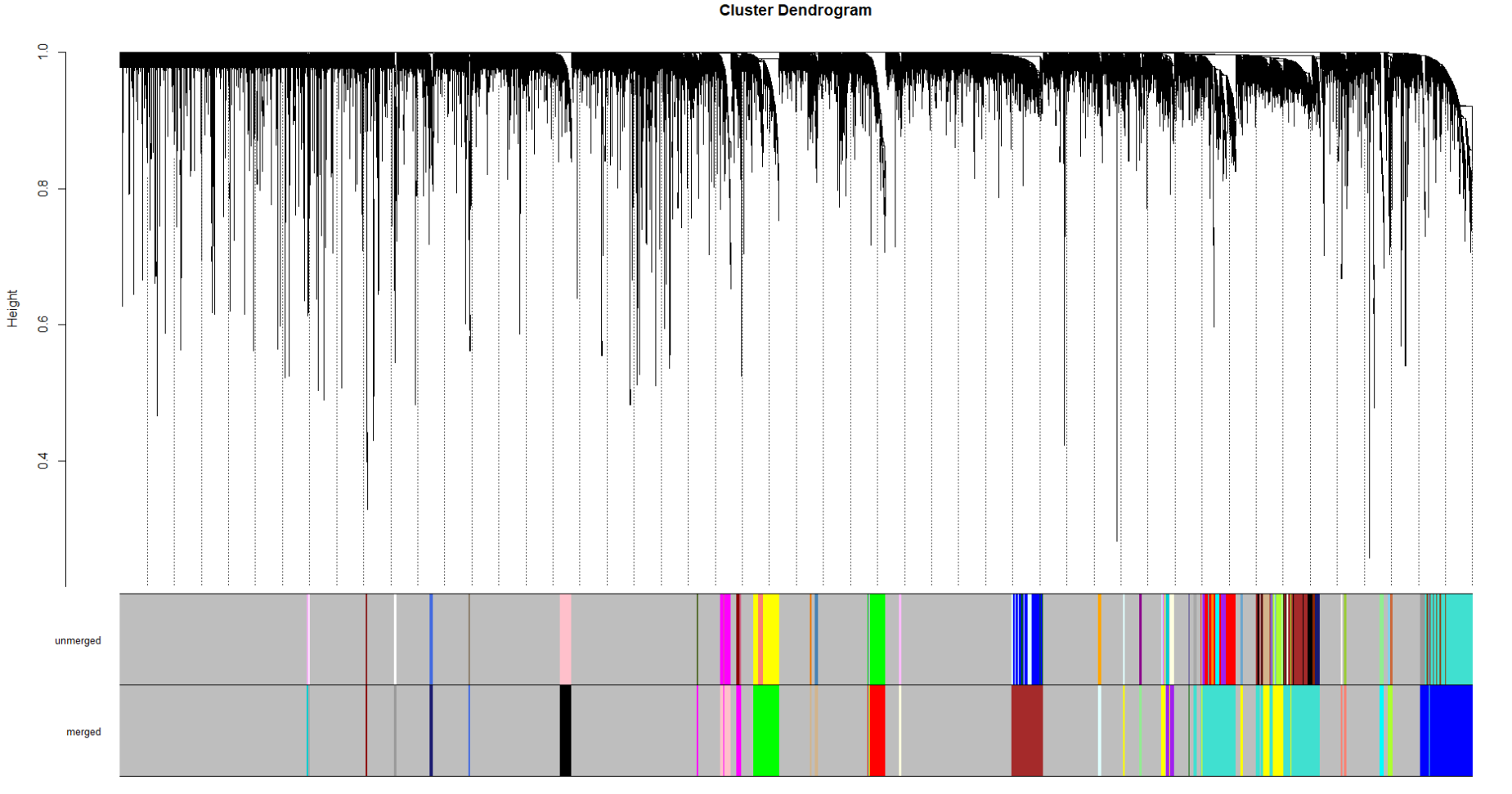
I'm having a trouble clustering TCGA GBM RNA data
One cluster is having massive genes and others have smaller.
I tried limma::removebatcheffect() but I think it's not working or I didn't do it correctly.
R version 4.3.0 (2023-04-21 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8 LC_CTYPE=English_United States.utf8 LC_MONETARY=English_United States.utf8 LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: Asia/Seoul
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] gridExtra_2.3 DESeq2_1.40.1 lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0
[6] dplyr_1.1.2 purrr_1.0.1 readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
[11] ggplot2_3.4.2 tidyverse_2.0.0 WGCNA_1.72-1 fastcluster_1.2.3 dynamicTreeCut_1.63-1
[16] SummarizedExperiment_1.30.2 Biobase_2.60.0 GenomicRanges_1.52.0 GenomeInfoDb_1.36.1 IRanges_2.34.1
[21] S4Vectors_0.38.1 BiocGenerics_0.46.0 MatrixGenerics_1.12.2 matrixStats_1.0.0 TCGAbiolinks_2.28.3
loaded via a namespace (and not attached):
[1] DBI_1.1.3 bitops_1.0-7 biomaRt_2.56.1 rlang_1.1.1 magrittr_2.0.3
[6] compiler_4.3.0 RSQLite_2.3.1 png_0.1-8 vctrs_0.6.3 rvest_1.0.3
[11] pkgconfig_2.0.3 crayon_1.5.2 fastmap_1.1.1 backports_1.4.1 dbplyr_2.3.2
[16] XVector_0.40.0 labeling_0.4.2 utf8_1.2.3 rmarkdown_2.22 tzdb_0.4.0
[21] preprocessCore_1.62.1 bit_4.0.5 xfun_0.39 zlibbioc_1.46.0 cachem_1.0.8
[26] jsonlite_1.8.5 progress_1.2.2 blob_1.2.4 DelayedArray_0.26.3 BiocParallel_1.34.2
[31] cluster_2.1.4 parallel_4.3.0 prettyunits_1.1.1 R6_2.5.1 stringi_1.7.12
[36] limma_3.56.2 rpart_4.1.19 Rcpp_1.0.10 iterators_1.0.14 knitr_1.43
[41] base64enc_0.1-3 downloader_0.4 R.utils_2.12.2 timechange_0.2.0 nnet_7.3-19
[46] Matrix_1.5-4.1 splines_4.3.0 tidyselect_1.2.0 rstudioapi_0.14 yaml_2.3.7
[51] doParallel_1.0.17 codetools_0.2-19 curl_5.0.1 lattice_0.21-8 plyr_1.8.8
[56] withr_2.5.0 KEGGREST_1.40.0 evaluate_0.21 foreign_0.8-84 survival_3.5-5
[61] BiocFileCache_2.8.0 xml2_1.3.4 Biostrings_2.68.1 pillar_1.9.0 BiocManager_1.30.21
[66] filelock_1.0.2 checkmate_2.2.0 foreach_1.5.2 generics_0.1.3 RCurl_1.98-1.12
[71] hms_1.1.3 munsell_0.5.0 scales_1.2.1 glue_1.6.2 Hmisc_5.1-0
[76] tools_4.3.0 data.table_1.14.8 locfit_1.5-9.8 XML_3.99-0.14 grid_4.3.0
[81] impute_1.74.1 AnnotationDbi_1.62.1 colorspace_2.1-0 GenomeInfoDbData_1.2.10 htmlTable_2.4.1
[86] Formula_1.2-5 cli_3.6.1 rappdirs_0.3.3 fansi_1.0.4 S4Arrays_1.0.4
[91] gtable_0.3.3 R.methodsS3_1.8.2 TCGAbiolinksGUI.data_1.20.0 digest_0.6.31 farver_2.1.1
[96] htmlwidgets_1.6.2 memoise_2.0.1 htmltools_0.5.5 R.oo_1.25.0 lifecycle_1.0.3
[101] httr_1.4.6 GO.db_3.17.0 bit64_4.0.5