Hi,
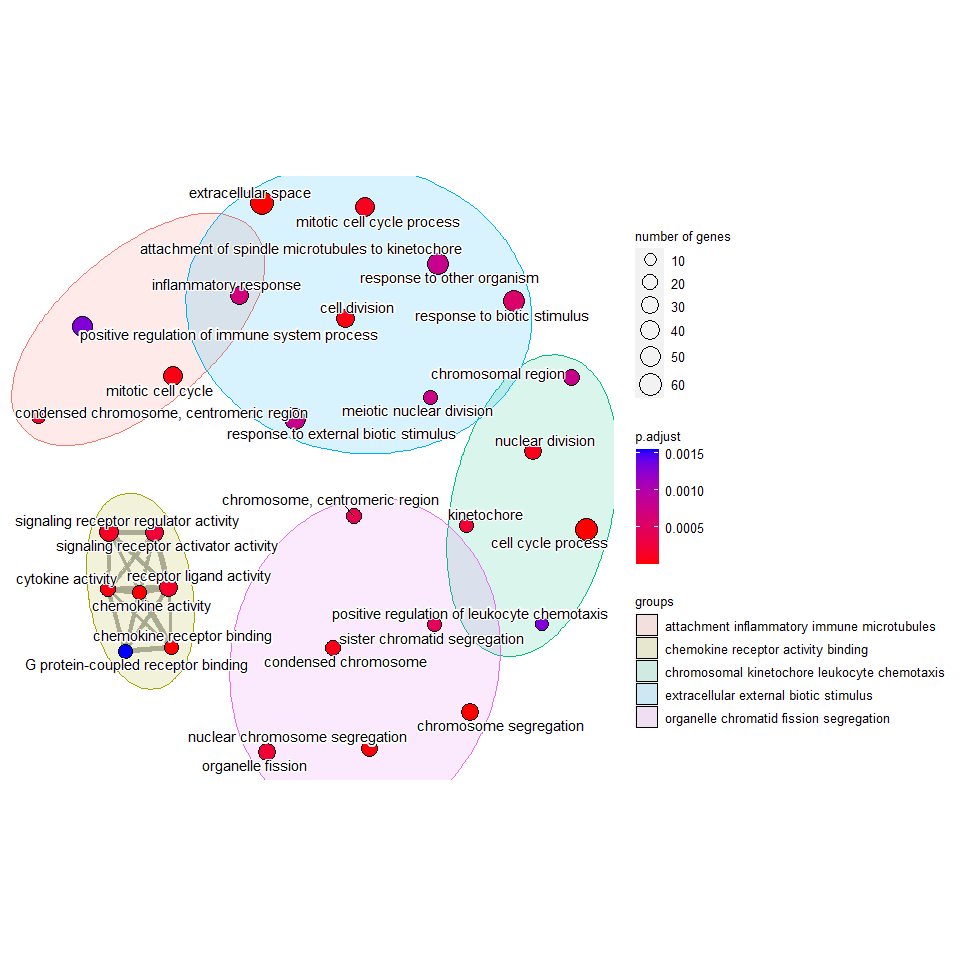
Using clusterprofiler an generated an emap plot from my enrichment data - would anyone be able to tell me where the grouping terms (bottom right) come from / how they are made? They look potentially like they are just picking common words?
Thanks, Matt


Absolutely brilliant, thanks for a definitive answer!
Thanks, Matt