I am trying to import coding sequences from biomaRt using the following code (simplified example):
cds_seq = getSequence(id = "NM_004974",
type = "refseq_mrna",
seqType = "coding",
mart = ensembl)
...and getting this output...
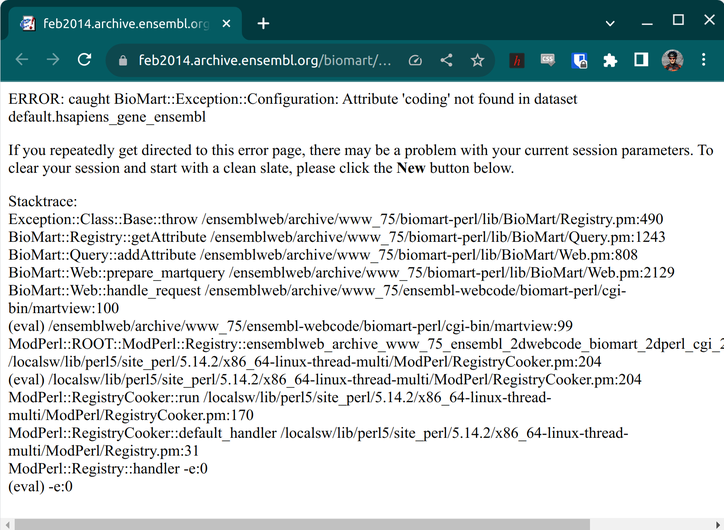
Error in biomaRt::getBM(attributes = c("hgnc_symbol", "ensembl_gene_id", :
Invalid attribute(s): coding
Please use the function 'listAttributes' to get valid attribute names
I ran the same code ~2 years ago and it worked fine. I've explored other Attributes via the "listAttributes()" function to see if the attribute name for coding sequence has been updated but I can't see to find anything close.
Note that I'm using an archived version of ensembl and wish to keep it that way so the code output doesn't change if any of the sequences of interest have been updated in newer versions. Not sure if this is relevant. Here's the command I used to import ensembl:
ensembl <- useEnsembl(biomart="ensembl",
dataset="hsapiens_gene_ensembl",
host="https://feb2014.archive.ensembl.org")
Please advise, and thank you!


This worked perfectly! I was able to retrieve the sequences of interest using
host="https://grch37.ensembl.org"within the useEnsembl call. Thanks to James and Mike (below) for your help, I really appreciate it.I should also point out that you can find this out by yourself by going to ensembl, clicking on the Biomart link, and then scrolling to the bottom and clicking on 'View in archive site`, which will bring up all the archives. There's one at the top that says
And hovering over the link gave me the host URI that I used.