Hi all, I really need some help. I am trying to download clinical data (xml files) for TCGA-BRCA project using TCGAbiolinks package and then merged all clinical files such as follow up, drug... using following command as suggested in TCGAbiolinks tutorial, which worked fine -
aux <- GDCprepare_clinic(query, clinical.info = i)
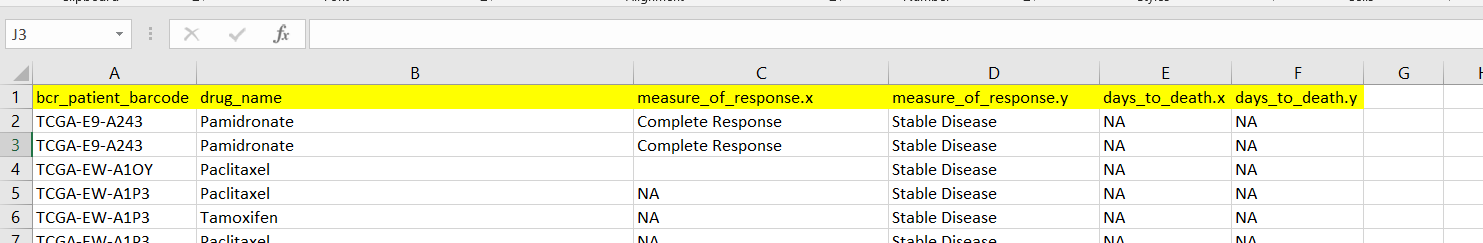
Now I got all the clinical xml files columns in one excel file (attached some cropped columns).
My question is there are two columns with similar information like measure of response.x and measure of response.y and both contain different treatment response information like complete response and other with stable disease. Which column should i consider for final study? for one patient what should i consider whether the patient completely responded to the treatment or the disease is stable?
Any help would be really appreciated. I'm literally stuck in my study.


pay and play will be your ticket to the fascinating world of online casinos! pay and play casino: https://onlinecasinoprofy.co.uk/pay-n-play-casinos/ is just a masterpiece site with all the right features! I recommend you to try it out, because I found a lot of great online slots and much more on this site!