I am working with a complex dataset design shown here: 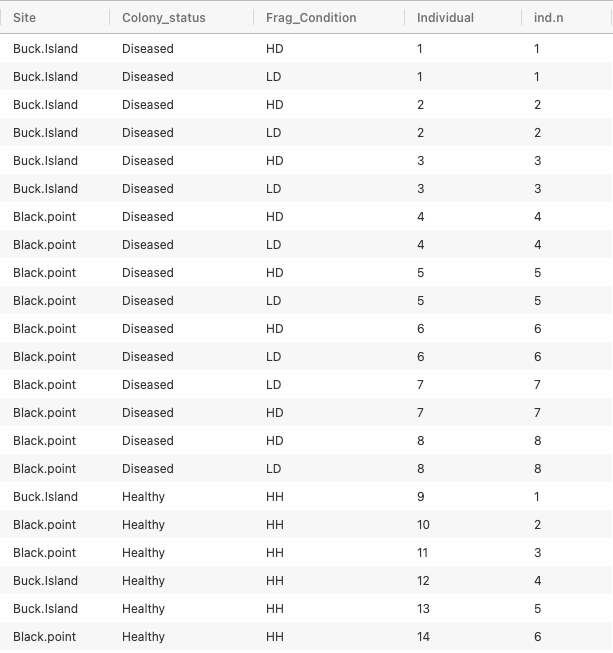
I have followed the DESeq2 manual, but have not been able to find an appropriate matrix model that tests for the effects of Frag_Condition while controlling for Site and Individual (Genotype) effects. I believe the error lies in the fact that the "HH" Frag_Conditions are unpaired and are nested within the "Healthy" Colony_status. Here is what I have tried:
countData <- countData
colData <- colData
# Convert colData columns to factors
colData$Site <- as.factor(colData$Site)
colData$Colony_status <- as.factor(colData$Colony_status)
colData$Frag_Condition <- as.factor(colData$Frag_Condition)
colData$Individual <- as.factor(colData$Individual)
colData$ind.n <- ave(as.factor(colData$Individual), colData$Colony_status, FUN = function(x) as.integer(factor(x)))
# Create model matrix
modelMatrix <- model.matrix(~Site + Colony_status:ind.n + Colony_status:Frag_Condition, colData)
# Remove all zero columns
all.zero <- apply(modelMatrix, 2, function(x) all(x==0))
all.zero
idx <- which(all.zero)
modelMatrix <- modelMatrix[,-idx]
modelMatrix
# Run DESeqDataSetFromMatrix
dds <- DESeqDataSetFromMatrix(countData = countData, colData = colData,design = modelMatrix)
# ERROR: Error in checkFullRank(modelMatrix) :
the model matrix is not full rank, so the model cannot be fit as specified.
One or more variables or interaction terms in the design formula are linear
combinations of the others and must be removed.
If anyone has any suggestions for how to fix my matrix design it would be greatly appreciated!!! I've been scratching my head for a while now, and at a loss.


Update: It seems like any form of trying to add Individual (Genotypic effects) to the model results in the full rank error. Moving forward with ~Site + Frag_Condition for now.
Would still welcome suggestions for improvement since I know this probably isn't the best model.
Yes, individual is inherently nested within Colony_status. For these sorts of things, as Mike says, you should seek local consultation, or try and post at a wider community such as biostars.org, though usually it takes some back and forth to find the right analysis strategy, arguing again for a local collaboration.