Hi everyone,
I'm getting an error when I try to run topGO using the Kolmogorov-Smirnov test and use the elim algorithm. I've set my gene list up so that it's a vector of all my genes with a calculated score to use for gene selection. It seems to run until it gets to the last level then throws an error. Not sure what I'm missing. Here's my code and the error message:
library(topGO)
library(org.Hs.eg.db)
de_matrix<-read.table("de_gene_matrix.txt", header=TRUE)
de_matrix<-na.omit(de_matrix) ## remove any rows with missing values
de_matrix$score<-ifelse((abs(de_matrix$log2FoldChange))>=1 & de_matrix$padj<0.05,
((abs(de_matrix$log2FoldChange))*(-log10(de_matrix$padj))),
0.00)
gene_list<-de_matrix$score
names(gene_list)<-de_matrix$symbol
GO2genes.BP <- annFUN.org(
whichOnto = 'BP',
feasibleGenes = NULL, ## no filtering of genes. all genes from GO are inlcuded.
mapping = 'org.Hs.eg.db',
ID = 'symbol')
selection<-function(score){
return(score>=1.3)
}
GOdata.BP <- new('topGOdata',
ontology = 'BP',
allGenes = gene_list,
annot = annFUN.GO2genes,
geneSel=selection,
GO2genes = GO2genes.BP,
nodeSize = 5)
go.bp.ks.elim=runTest(GOdata.BP, algorithm='elim', statistic='ks')
-- Elim Algorithm --
the algorithm is scoring 8428 nontrivial nodes
parameters:
test statistic: ks
cutOff: 0.01
score order: increasing
Level 18: 2 nodes to be scored (0 eliminated genes)
Level 17: 12 nodes to be scored (0 eliminated genes)
Level 16: 40 nodes to be scored (0 eliminated genes)
Level 15: 71 nodes to be scored (43 eliminated genes)
Level 14: 117 nodes to be scored (85 eliminated genes)
Level 13: 243 nodes to be scored (197 eliminated genes)
Level 12: 463 nodes to be scored (498 eliminated genes)
Level 11: 802 nodes to be scored (853 eliminated genes)
Level 10: 1093 nodes to be scored (2493 eliminated genes)
Level 9: 1229 nodes to be scored (5036 eliminated genes)
Level 8: 1296 nodes to be scored (5754 eliminated genes)
Level 7: 1173 nodes to be scored (6587 eliminated genes)
Level 6: 932 nodes to be scored (8074 eliminated genes)
Level 5: 545 nodes to be scored (9262 eliminated genes)
Level 4: 289 nodes to be scored (9991 eliminated genes)
Level 3: 102 nodes to be scored (10990 eliminated genes)
Level 2: 18 nodes to be scored (11349 eliminated genes)
Level 1: 1 nodes to be scored (11349 eliminated genes)
Error in if (termSig <= adjs.cutOff) { :
missing value where TRUE/FALSE needed


Thanks for you response. Could you elaborate on how to step through this? I ran the debug code you gave above and hit 'return' to step through to the
for(i in nodeLevel$noOfLevels:1)step. When I hit 'c' at any point here, it runs through the rest of the code and gives the error message. It will step through if I continue to hit 'return' but it goes through each GO term and takes forever. I guess my question actually is "is there a way to speed this up?" Is it possible to step through each level for i where it stops after each iteration or do I have to just live with holding the return key for an hour or so? I apologize in advance for the ignorance of my question. I've been using R for a long time but I've never had to debug like this.Oh, my bad. It was my recollection that you could just hit 'c' to jump to the next iteration of the loop, but I think that's wrong. Unfortunately it looks like you will have to just iterate through them all...
@james-w-macdonald-5106 Again, thanks for your response. I ran through the whole list of terms. At level i = 1L there is only one node to test, GO:0001850 which is the top parent term in biological process, i.e. biological process. When the function
termSig <- runTest(group.test)is run, it produces a value of "NA_real_" for termSig which is causing it to fail. But I can't figure out why. There doesn't seem to be any issues that I can tell with any of the parameters.I know without having eyes on my actual data it would be difficult to diagnose but do you know of any particular reason offhand that it could be behaving like this? Is there a way to fix it? Apologies up front for asking the impossible. Here is an image of the envrionment at i=1 if that helps at all.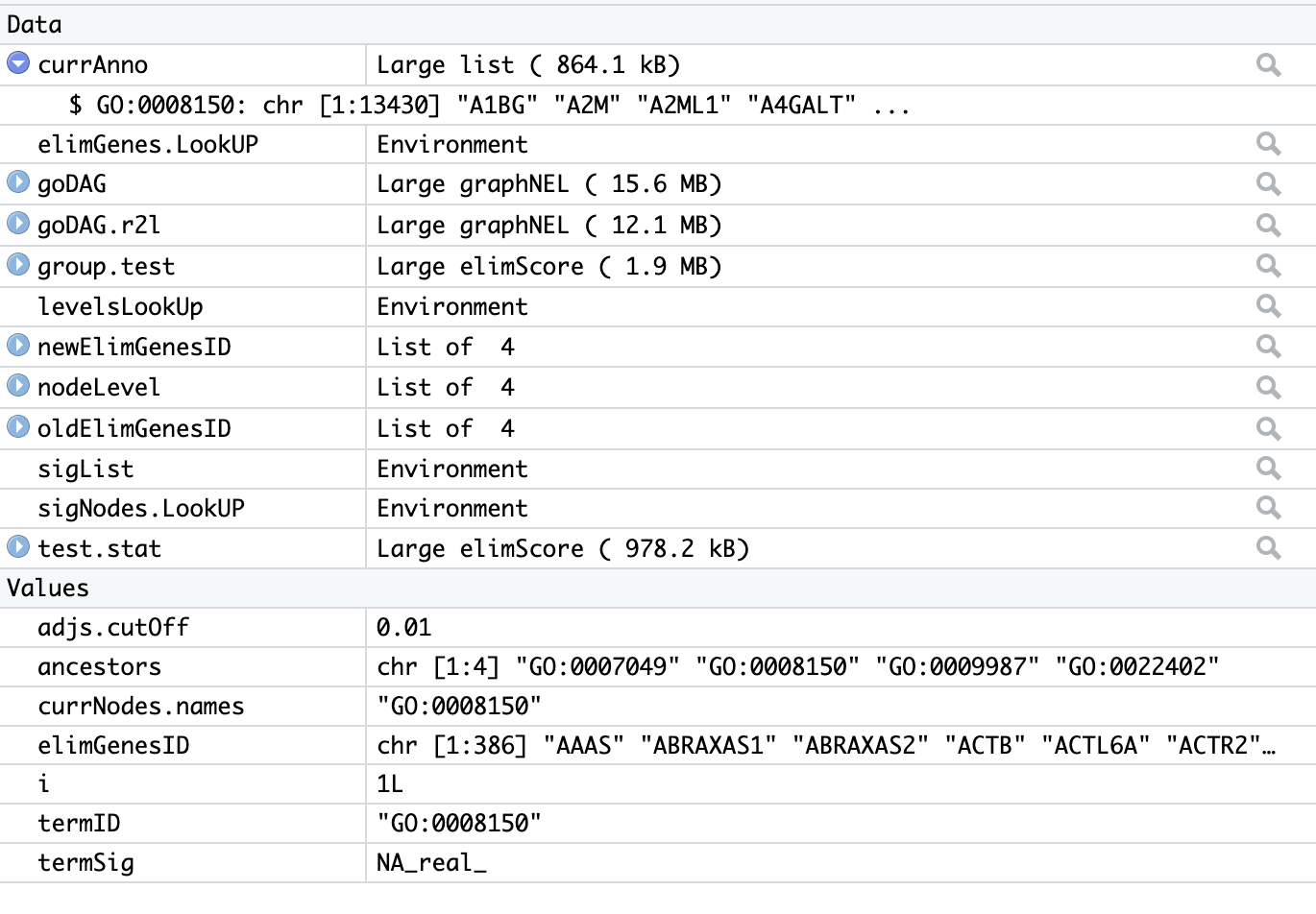
I tried using all the same parameters except I used the values for adjusted p value in the selection function and it worked using the K-S test and the elim algorithm. However, if I calculate the -log10 of the adjusted p value and use that, it fails with the same error message. So it clearly is a problem with the numbers used for selection. I'm at a loss as to why. Anyway, I very much appreciate your input. If you have ideas on what can be done to work around this, please let me know.