Hello
I am looking for help with the analysis of bulk RNA-seq data. I have an analysis in mind, but I would like to verify its correctness.
Experiment:
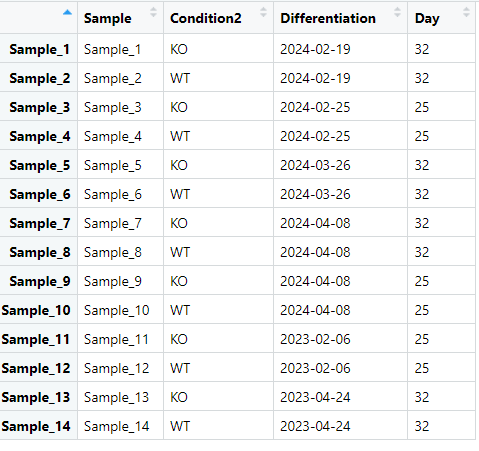
We differentiate stem cells into embryonic bodies. These embryonic bodies produce progenitor cells that are further differentiated into a certain cell type. These progenitors can be harvested at two time points: day 18 or day 25 after which the second differentiation protocol starts. These "second differentiated cells" were sent for bulk RNA sequencing.
We have both wild type cells and knock-out (KO) samples. Our primary question is which genes are differentially expressed between wild type and KO samples. A secondary question is whether there are differences between day 18 and day 25.
A PCA plot shows that "Differentiation" (one embryonic body) is a significant source of variation. This variation is captured in PC1. I think it is correct to include "Differentiation" in my model, but I believe it is not possible with fixed effects since there is not always a day 18 and day 25 present in each "Differentiation"
I would like to include this in my analysis, but I believe it is not possible with fixed effects since there is not always a day 18 and day 25 in each "Differentiation".
My Design:
Inspired by section 7.6 of "A Guide to Creating Design Matrices for Gene Expression Experiments," (great resource by the way!) my idea was to use limma with a design matrix "~Day + Condition2" and include "Differentiation" as a random effect (block becomes "Differentiation" in duplicateCorrelation and lmFit), because the blocks are not complete (not every Day and Condition is in every differentiation). The last parameter in my design is then my effect of primary interest.
Is this a correct way to analyze this data?
Thank you very much for your time and help.