I performed two RNA-Seq experiments in exactly the same way, with the only difference being the studied cell type (mouse cortical neurons vs sensory DRG neurons). Everything about these experiments suggests they went really well, and the enriched genesets make a lot of sense.
About the attached volcano plots: Do I need to be concerned about the fact that the p value range is smaller in one experiment ? If the reason is in the underlying biology (and not a technical issue), is the explanation that a higher variability of individual measurements is sometimes observed ?

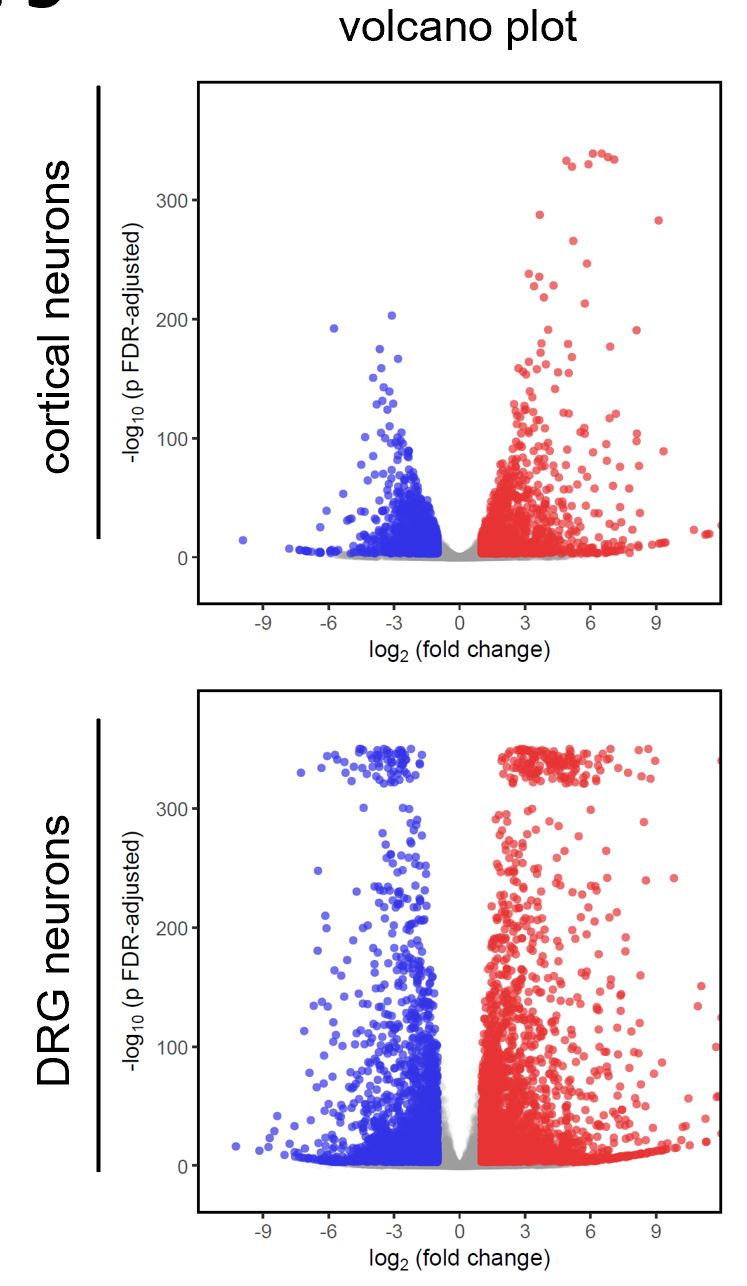

Thanks - all I wanted to hear (and I believe that is what you are saying) is that it is not concerning if p value ranges between two very similar experiments differ in the way shown. I am not trying to figure out the reason because I wouldn't know what to learn from it.