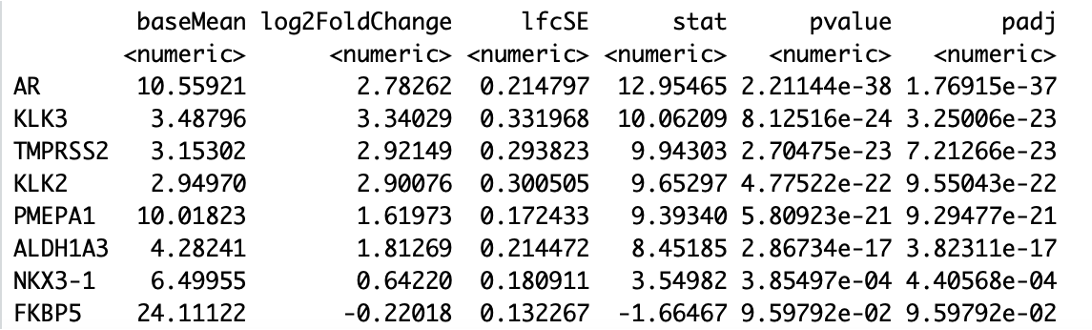
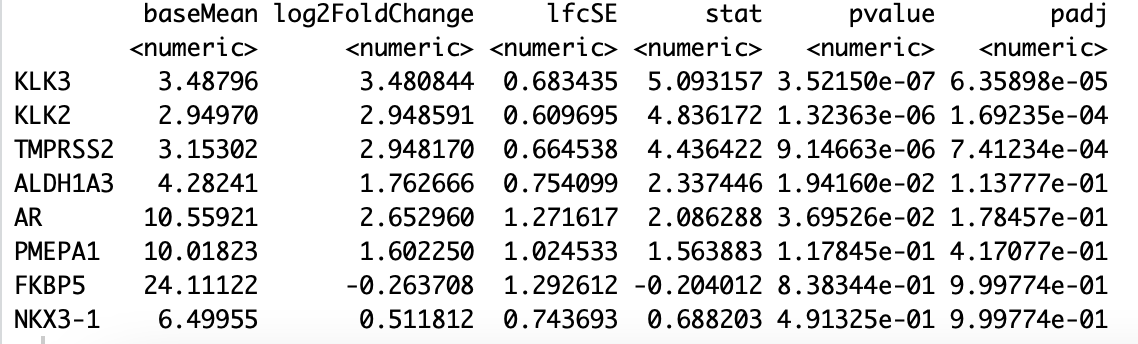
I ran rstudio on my college's server last week, and DESeq2 results looked good to me. However, I forgot to save the environment, I just took a screenshot of DESeq results before I log out. after one week, I opened the R file, and re-ran it, I got the different results. I compared the current results with the screenshot I saved, baseMean are the same, others (log2FoldChange, lfcSE, state, value, padj) are different. for example, log2FC changed a little bit, however, pvalue/padj became much larger. both results are sorted by padj, and only display selected genes. Do anyone know the reason? I"m attaching the comparisons as follows. thank you.
before (good results):
now (bad results):