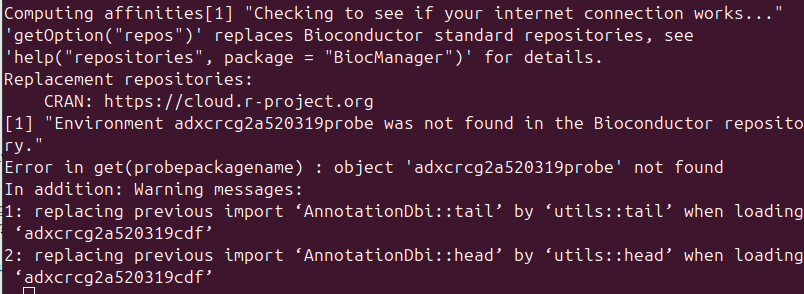
Hi, This is my first time working with AffyMatrix .CEL files. I was hoping to extract the probeset gene expression data for the E-MTAB-990 dataset which uses a custom Affymetrix A-AFFY-101. My Code follows the standard pipeline of downloading all files from ArrayExpress and then running gcrma. The problem stems from the error which says "Error in get(probepackagename) : object 'adxcrcg2a520319probe' not found." I have tried looking at makeProbePackage however I'm not sure how to create the "probe_tab" file for A-AFFY-101. The CDF environment is created fine however the last line calling grcma line throws the error.
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("makecdfenv")
BiocManager::install("affy")
BiocManager::install("ArrayExpress")
BiocManager::install("limma")
BiocManager::install("AnnotationDbi")
BiocManager::install("gcrma")
library("ArrayExpress")
library("makecdfenv")
library("affy")
library("AnnotationDbi")
library("limma")
library("gcrma")
setwd('FOLDER_WITH_CDF_FILE')
make.cdf.package("ADXCRCG2a520319.cdf", "adxcrcg2a520319", species = "Homo sapiens", compress = TRUE)
install.packages("adxcrcg2a520319/", repos = NULL, type = "source")
library("adxcrcg2a520319")
mexp1422 = getAE("E-MTAB-990", type = "full")
rawset <- ReadAffy(cdfname="adxcrcg2a520319")
eset <- gcrma(rawset,type="affinities",fast=FALSE,affinity.source="reference",normalize=TRUE)