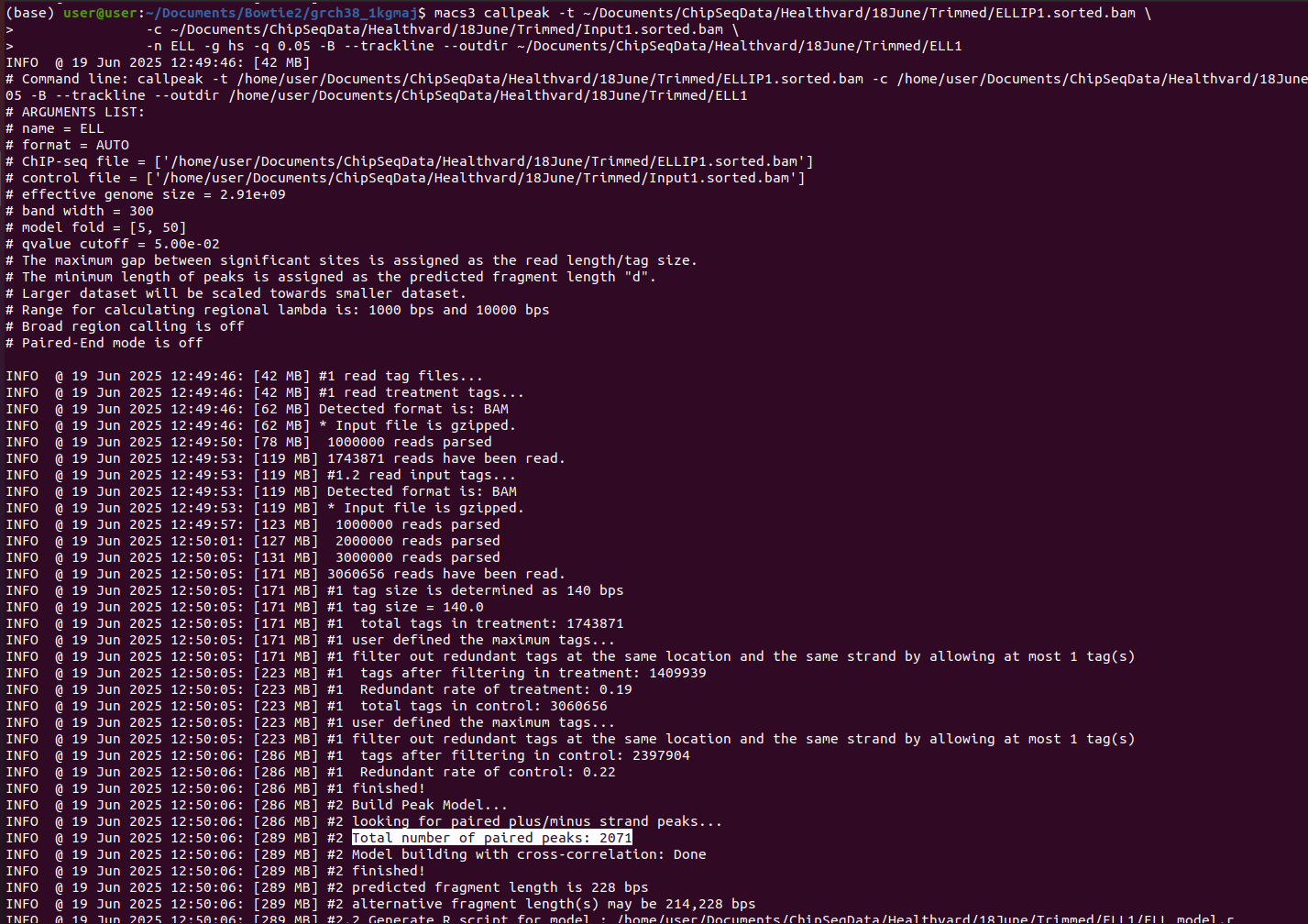
``` Some help needed regarding Chip-seq analysis. I use MACS3 to call the peaks. Earlier, I had used a Chip-seq sample data from the internet and obtained around 60,000 peaks/sorted.bam file from the pipeline that I use.
But now I am getting around 50 peaks only in the sample which we prepared. The input IP control is also there. How do we know whether there is a problem in the sample or the software/pipeline/MACS3 ?