I am using HiCDCPlus to normalize intact Hi-C data (MNase-based, from ENCODE: ENCFF091YKP, HUVEC cells, 10kb resolution, hg38). I'm experiencing poor z-score distribution compared to in-situ Hi-C data, despite extensive parameter tuning, and I hope you can provide guidance.
Problem Description
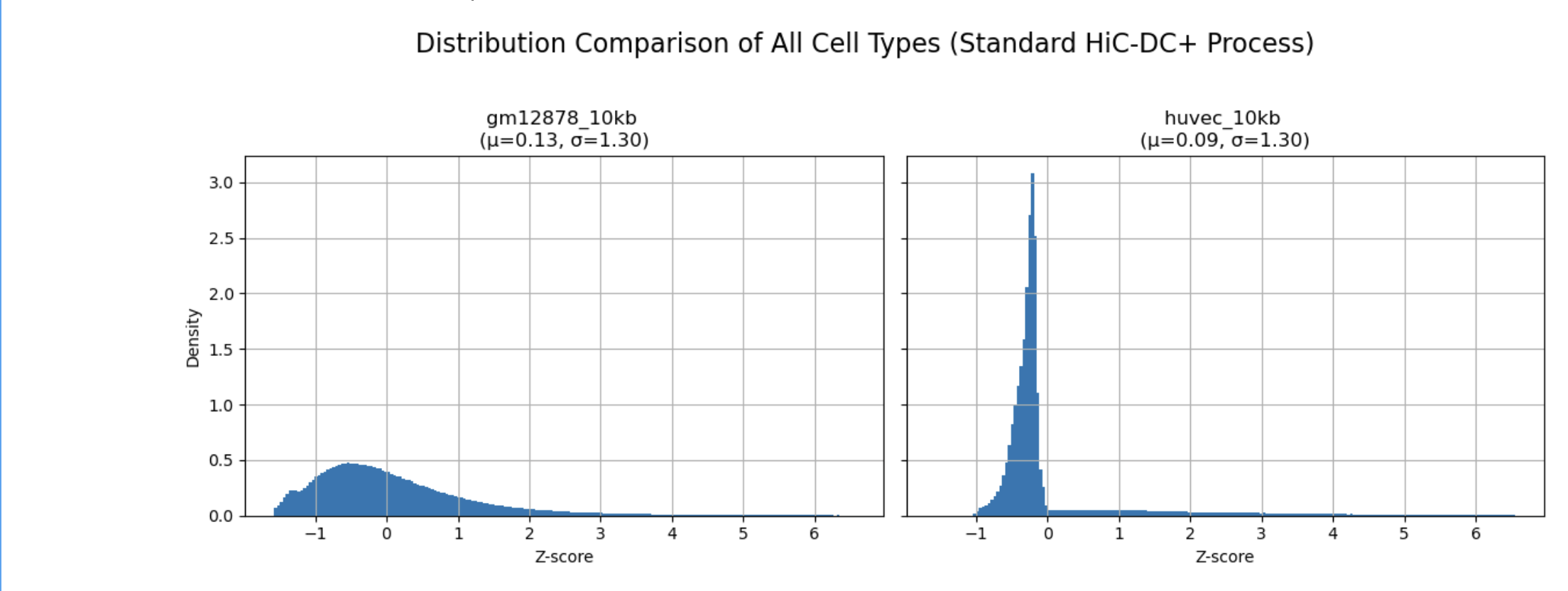
When using feature_type="RE-agnostic" for intact Hi-C data, the z-score distribution shows a sharp peak at zero with rapid decay, unlike the smooth bell-shaped distribution observed with RE-based in situ Hi-C data. I don't know what I should do to increase the preprocessing performance.
Attempts that didn't help
- Increasing ssize from 0.01 to 0.9: sigma only changed from 1.35 to 1.30
- Adding mappability bigWig: minimal improvement
- Adding nuclease cleavage frequency: sigma improved to 1.30 but distribution shape unchanged
- Filtering gc=map=0 interactions: no significant change ```r # My code construct_features( feature_type = "RE-agnostic", wg_file = "k50.Umap.MultiTrackMappability.bw" )
gi_list <- add_1D_features(gi_list, mnase_cleavage_data) gi_list <- expand_1D_features(gi_list) gi_list <- gi_list[mcols(gi_list)$mnase_cleavage != 0]
gi_list <- HiCDCPlus_parallel( gi_list, covariates = c("gc", "map", "mnase_cleavage"), model_distribution = "nb", ssize = 0.9, df = 6, Dmin = 0, Dmax = 2e6 )
Session info
sessionInfo()
```