Hello,
I am performing differential expression analysis using DESeq2 on human RNA-seq data with 3 donors per condition.
I am confused by the behavior of lfcThreshold in results(). When I specify lfcThreshold instead of post-hoc LFC filtering, some genes with very low expression and biologically negligible change are still reported as significant and would pass into downstream enrichment analysis.
Below is an example.
# Without applying lfcThreshold
summary(results(dds_t1,
name = "Dose_4Gy_vs_0Gy",
alpha = 0.05))
This returns:
- out of 18591 with nonzero total read count
- adjusted p-value < 0.05
- LFC > 0 (up) : 530, 2.9%
- LFC < 0 (down) : 739, 4%
# With lfcThreshold
summary(results(dds_t1,
name = "Dose_4Gy_vs_0Gy",
alpha = 0.05,
lfcThreshold = 0.3))
This returns:
- out of 18591 with nonzero total read count
- adjusted p-value < 0.05
- LFC > 0.30 (up) : 176, 0.95%
- LFC < -0.30 (down) : 147, 0.79%
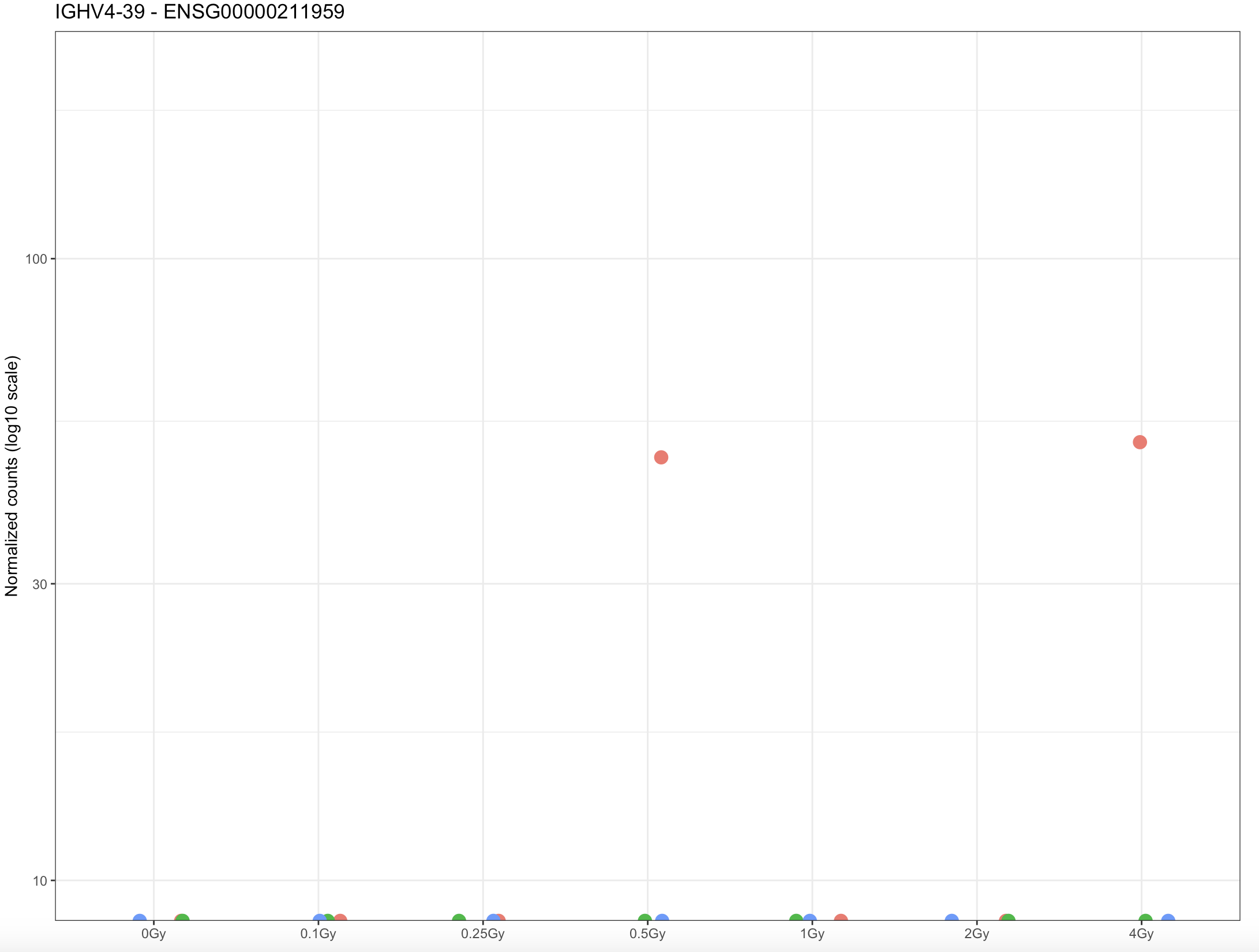
In the second case, with lfcThreshold, I observed that the gene ENSG00000211959 is reported as differentially expressed with:
padj = 0.04831358
log2FoldChange = 18.78
a shrunken LFC approx. 0.0047
However, this gene is barely expressed across samples (as shown in the expression plot), and it does not appear as significant when lfcThreshold is not used. I observe similar behavior for other lowly expressed genes.
Questions:
Is this behavior expected when using
lfcThreshold, particularly for lowly expressed genes?Would it be appropriate to apply an additional post-hoc filter, for example, on shrunken LFC, when using
lfcThreshold, especially before enrichment analysis?What would be the appropriate way to identify meaningful/True DE genes to report them and pass them to enrichment to put them into biological context?
Thanks in advance for any recommendation and clarification.


Thank you very much for your answer.
Actually, this is after pre-filtering
would you recommend a stricter filtering?
To note, I only observed this behavior after applying the
lfcThreshold. These lowly expressed genes were not detected as significant without thelfcThreshold. I am using thelfcThresholdonly because I wanted to get rid of DE genes with low effect size instead of doing a post-hoc filtering on the LFC.it looks above like you have a gene with two non-zero count samples.
Also, yes you can use a stricter filter. You cannot estimate the LFC when most of the samples do not detect the gene at all.
Jumping in on this - all this started because we want to "do it right", i.e. avoiding the post-hoc filtering on the logFC (which is still way too widely used).
So we set out to go with
lfcThreshold, and at the same time, for the sake of keeping all genes in (relevant if we want to compare to other datasets), no filtering done up ahead.We noticed the fact we obtained very high logFC values, that then get (correctly) shrunken. Yet they do somehow get low p-values, "even if the variability is so high". So the unshrunken logFC goes through the parameter value we set. Probably it stems from the surprise effect I get from seeing those genes get a very significant p-value.
We were wondering if it is possible (and statistically kosher) to go with re-adding a round of post hoc logFC threshold, "since we anyway tested against the more stringent null hypothesis".
... and possibly: the choice of lfcThreshold does influence the features that get used for testing via independent filtering - a p-value is there for all, but the adjustment is not performed in some cases.
Thank you for the suggestions!
Yes, I find that the LFC shrinkage and its corresponding stat
svalueif you use lfcShrink with a threshold often returns more reliable gene sets.I think adding the use of
lfcThresholdmakes sense. I wouldn't recommend post-hoc LFC thresholding outside of whatresults()function provides, as the adjusted p-value then doesn't correspond to the final selected set.Yes, so I would think that you've changed the order of
padjhere but not p-value. I don't think use oflfcThreshold> 0 can decrease (more significant) the p-value.