Hi all,
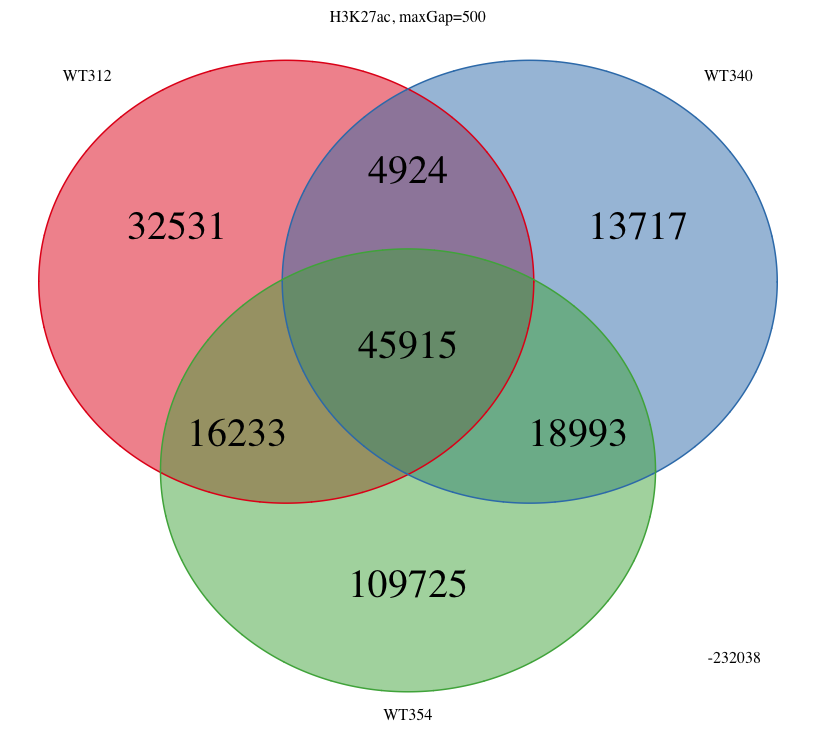
I was wondering if there is anyway to get ChIPpeakAnno to report the total number of covered bases (as opposed to the number of overlapping regions) when making use of makeVennDiagram()? Is this possible? Would it be difficult to add this as a feature?
Thanks,
Brad