I am getting drastically different result when analysing a dataset using different version of DESeq2.
I'm repeating DE analysis I did last year, with a previous version DESeq2 - specifically, DESeq2 1.4.5, under R3.1 Spring Dance - and I am seeing completely different results with the current version, which is is DESeq2 1.8.1 under R 3.2.1 World-Famous Austronaut.
I'm analysing differential expression in a microbiome dataset. Wiht 1.4.5 I get lots of differental expression and a trend towards negative expression. I get a vastly different result with the new DESeq version - significantly less DE and no trend. Biologically, I would expect the results to conform to the old version with plenty of neg. I've attached MA plots to show an example - same dataset here, DEseq 1.8.1 on the left and 1.4.5 on the right.
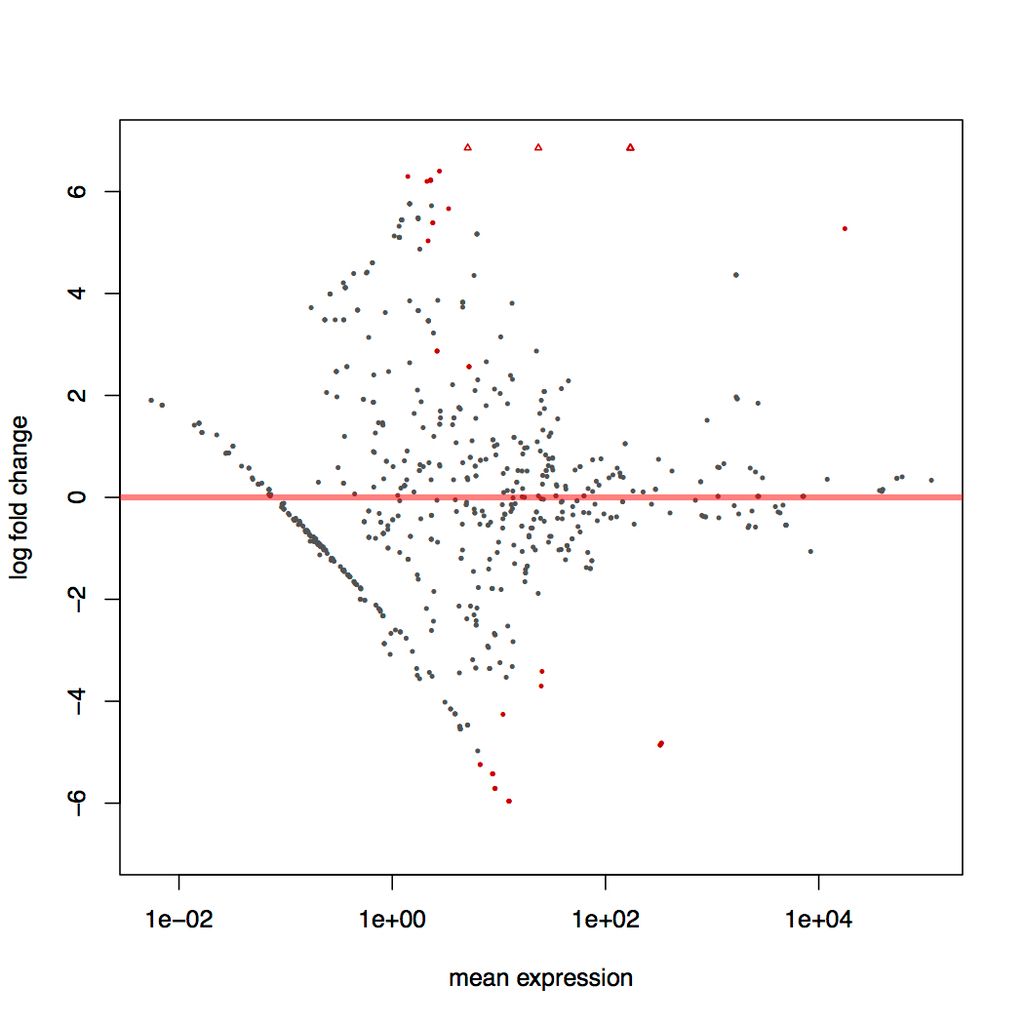

There are a lot of instances in my data where - say, if I have 8 'positive' and 8 'negative' samples for my condition, one positive sample may have signifciant DE of some OTU but no other samples have any of that OTU. This usually results in a non-significant p value under the old version, and a p value of 1 (and NA logFC) for the new version. However, most of the significant DE that I 'lose' isn't from cases like this, though there are still plenty of instances where e.g. half the samples have no count of some OTU.
I'm using phyloseq upstream of the DESeq2 calls to build my dataframes, if that matters at all. OTU picking, upstream, was done with QIIME. I've debugged my pipeline and am certain it's DESeq2 responsible.
I've figured out that the discrepancy happens between DESeq2 1.6.3 and DESeq2 1.4.5 - I originally suspected the changes at 1.7.32, but - 1.6.3 gives me the 'new' low DE results. Looking at the changelog between 1.4.5 and 1.6.3, there's Cook's Distance stuff, mostly, that's been changed?
Any clues or ideas? I didn't expect a version update to make such a drastic change in DE results. Thank you in advance.

