I wouldn't really want it to be a data.frame, given the one-to-many mappings, but that's up to you.
> gos <- paste0("GO:", sprintf("%07d", c(10468,19222,60255,9889,10556,31326,2000112)))
> gos
[1] "GO:0010468" "GO:0019222" "GO:0060255" "GO:0009889" "GO:0010556"
[6] "GO:0031326" "GO:2000112"
> z <- mapIds(org.Pf.plasmo.db, gos, "ORF", "GOALL", multiVals = "list")
'select()' returned 1:many mapping between keys and columns
> sapply(z, length)
GO:0010468 GO:0019222 GO:0060255 GO:0009889 GO:0010556 GO:0031326 GO:2000112
161 183 180 150 149 150 149
> lapply(z, head)
$`GO:0010468`
[1] "PF3D7_0109600" "PF3D7_0110800" "PF3D7_0111800" "PF3D7_0204600"
[5] "PF3D7_0209700" "PF3D7_0212300"
$`GO:0019222`
[1] "PF3D7_0109600" "PF3D7_0110800" "PF3D7_0111800" "PF3D7_0204600"
[5] "PF3D7_0205900" "PF3D7_0209700"
$`GO:0060255`
[1] "PF3D7_0109600" "PF3D7_0110800" "PF3D7_0111800" "PF3D7_0204600"
[5] "PF3D7_0205900" "PF3D7_0209700"
$`GO:0009889`
[1] "PF3D7_0109600" "PF3D7_0110800" "PF3D7_0111800" "PF3D7_0204600"
[5] "PF3D7_0209700" "PF3D7_0212300"
$`GO:0010556`
[1] "PF3D7_0109600" "PF3D7_0110800" "PF3D7_0111800" "PF3D7_0204600"
[5] "PF3D7_0209700" "PF3D7_0212300"
$`GO:0031326`
[1] "PF3D7_0109600" "PF3D7_0110800" "PF3D7_0111800" "PF3D7_0204600"
[5] "PF3D7_0209700" "PF3D7_0212300"
$`GO:2000112`
[1] "PF3D7_0109600" "PF3D7_0110800" "PF3D7_0111800" "PF3D7_0204600"
[5] "PF3D7_0209700" "PF3D7_0212300"
If you want a data.frame it's easy enough
> zz <- data.frame(GO = rep(names(z), sapply(z, length)), ORF = do.call(c, z))
> head(zz)
GO ORF
GO:00104681 GO:0010468 PF3D7_0109600
GO:00104682 GO:0010468 PF3D7_0110800
GO:00104683 GO:0010468 PF3D7_0111800
GO:00104684 GO:0010468 PF3D7_0204600
GO:00104685 GO:0010468 PF3D7_0209700
GO:00104686 GO:0010468 PF3D7_0212300
The number of genes I get is different from what you get, probably because you have a smaller universe. You can easily filter on genes that were in your original universe.

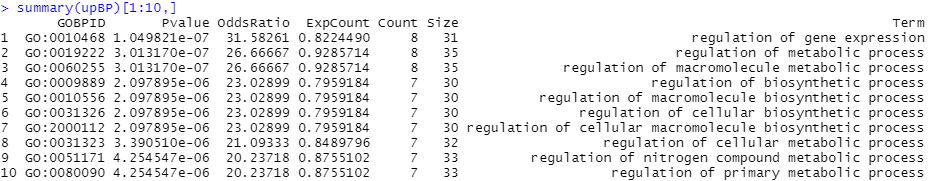

I am afraid that in this case ZZ returns the results from the oeginal database "org.Pf.plasmo.db" (all genes included) i want to list only those in upBP