I asked on the Slack channel
Any specific reason why the downstream is set to 200 in the GenomicFeatures::promoters function?
Herve Pages pointed out
This is just a default value that was set to be consistent with GenomicFeatures::getPromoterSeq(). Paul Shannon is the original author of getPromoterSeq(). I don't know if Paul is on this slack but if you ask on our support site you might get a response from him.
I would like to know if there is more to it

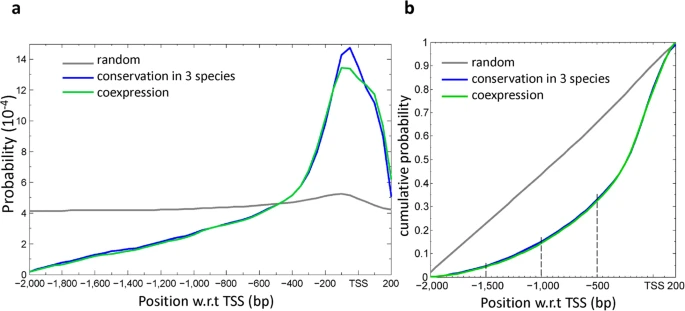
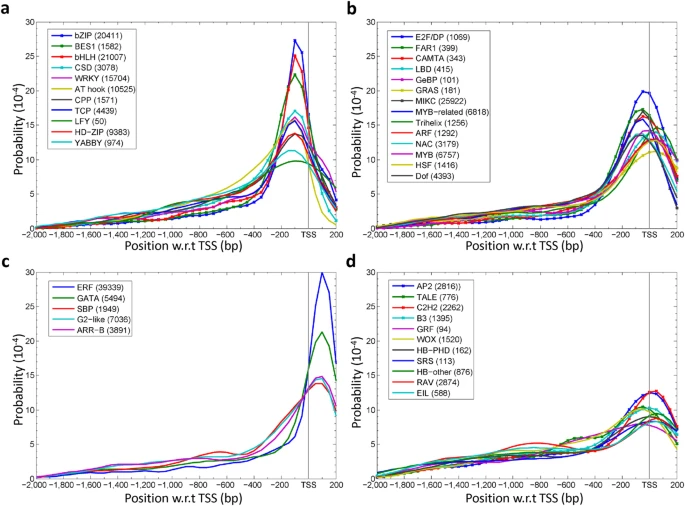

I just asked on GitHub too: https://github.com/Bioconductor/GenomicFeatures/issues/35