Hello,
I used TxDb.Hsapiens.UCSC.hg38.knownGene/GenomicFeatures to retrieve gene promoters and other genomic features.
here is code:
library('TxDb.Hsapiens.UCSC.hg38.knownGene')
txdb <- TxDb.Hsapiens.UCSC.hg38.knownGene
PR <- promoters(txdb, upstream=2000, downstream=0)
but when I take a look at the PR results:
it is quite weird. As some of promoters does not belong to any genes.
Now the fact that some transcripts are not associated with any gene is just the way things are in the GENCODE V38 track from the UCSC folks, which is the track that TxDb.Hsapiens.UCSC.hg38.knownGene is based on.
Thanks a lot.
I also get another issue:
I used threeUTRsByTranscript(txdb, use.names=TRUE)) to extract 3'UTR of genes, but the start and end coordinate of the 3'UTR highlighted in blue is the same, which looks weird.
for other genomic features, such as exon, I also get the same problem:
Does anyone like to explain this phenomenon?
Screen Shot 2021-12-12 at 1.00.48 PM
ok ok let's slow down and take the time to discuss the basics:
This is a different question. Your original question has been answered. If you're satisfied with the answer, please mark it as an accepted answer. If you're not, then please tell us why here. For any new question, please start a new thread.
Show the exact commands you use as well as their output. This should all be done in markdown. No need to upload screenshots that show the cropped version of some random output and with your drawings on top of it.
Avoid the use of things like "looks weird" or "explain this phenomenon". This is vague and subjective. Instead describe (with words) exactly and precisely what the problem is.
Looking forward to answer your next question in a different thread.
threeUTRsByTranscript(txdb, use.names=TRUE))to extract 3'UTR of genes, but the start and end coordinate of the 3'UTR highlighted in blue is the same, which looks weird.for other genomic features, such as exon, I also get the same problem: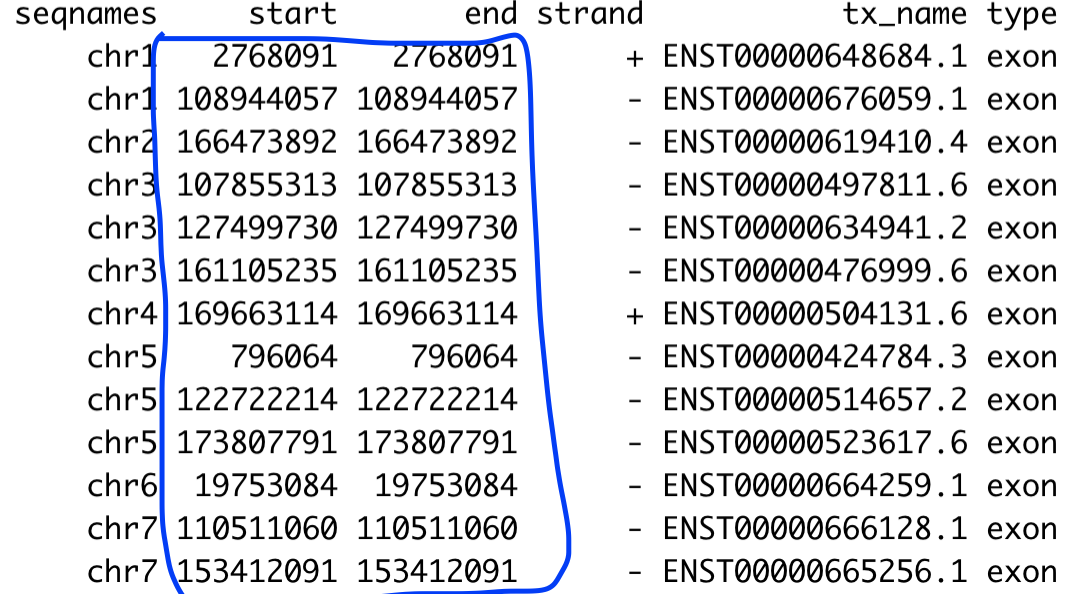
Does anyone like to explain this phenomenon? Screen Shot 2021-12-12 at 1.00.48 PM
ok ok let's slow down and take the time to discuss the basics:
This is a different question. Your original question has been answered. If you're satisfied with the answer, please mark it as an accepted answer. If you're not, then please tell us why here. For any new question, please start a new thread.
Show the exact commands you use as well as their output. This should all be done in markdown. No need to upload screenshots that show the cropped version of some random output and with your drawings on top of it.
Avoid the use of things like "looks weird" or "explain this phenomenon". This is vague and subjective. Instead describe (with words) exactly and precisely what the problem is.
Looking forward to answer your next question in a different thread.
Thanks,
H.