Entering edit mode
Using the Gviz library, I would like to transform data on the fly when plotting. This should be possible using the DataTrack arguments window, windowSize, and aggregation. However, it seems that the aggregation argument is set to "mean" and cannot be changed.
library(Gviz)
set.seed(0)
gr <- GRanges(seqnames = "chrI", ranges = IRanges(start = 1:100, width = 1), score = sample(x = 1:100, size = 100, replace = TRUE))
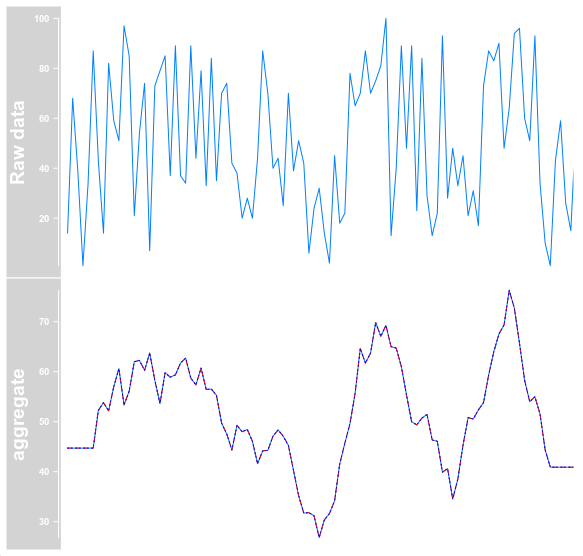
dt <- DataTrack(range = gr, type = "l", name = "Raw data")
dt_mean <- DataTrack(range = gr, type = "l", window = -1, windowSize = 11, aggregation = "mean", name = "aggregate")
dt_max <- DataTrack(range = gr, type = "l", window = -1, windowSize = 11, aggregation = "max", col = "red", lty = "dashed")
dt_min <- DataTrack(range = gr, type = "l", window = -1, windowSize = 11, aggregation = "min", col = "blue", lty = "dotted")
plotTracks(trackList = list(dt, OverlayTrack(trackList = list(dt_mean, dt_max, dt_min))), chromosome = "chrI")
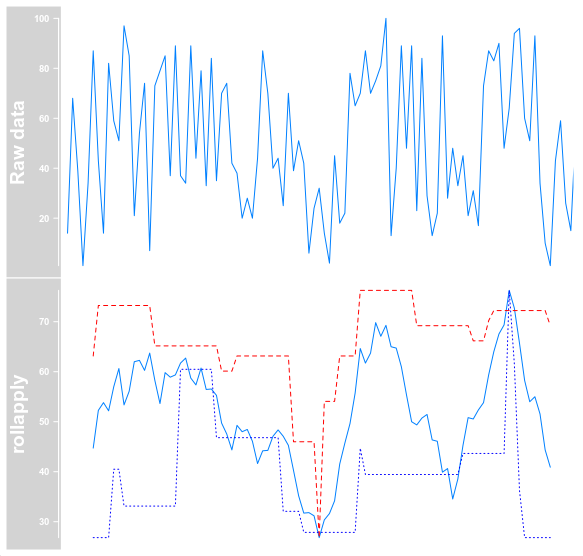
When I calculate min and max manually, I get the expected result.
library(zoo)
gr_mean <- gr
gr_mean$score <- rollapply(data = gr_mean$score, width = 11, FUN = mean, fill = NA)
dt_manual_mean <- DataTrack(range = gr_mean, type = "l", name = "rollapply")
gr_max <- gr
gr_max$score <- rollapply(data = gr_max$score, width = 11, FUN = max, fill = NA)
dt_manual_max <- DataTrack(range = gr_max, type = "l", col = "red", lty = "dashed")
gr_min <- gr
gr_min$score <- rollapply(data = gr_min$score, width = 11, FUN = min, fill = NA)
dt_manual_min <- DataTrack(range = gr_min, type = "l", col = "blue", lty = "dotted")
plotTracks(trackList = list(dt, OverlayTrack(list(dt_manual_mean, dt_manual_max, dt_manual_min))), chromosome = "chrI")
Am I missing something about how to correctly use the aggregation argument or is this a bug?
sessionInfo( )
# attached base packages:
# [1] grid stats4 stats graphics grDevices utils datasets methods base
# other attached packages:
# [1] zoo_1.8-9 Gviz_1.38.0 GenomicRanges_1.46.1 GenomeInfoDb_1.30.0 IRanges_2.28.0 S4Vectors_0.32.3 BiocGenerics_0.40.0