Hello everyone, I'm trying to analyse microarray data from Affymetrix. I am analysing a study in GEO database with a number:GSE49893. Unfortunately I have stumbled upon a problem. When using a 'alias2SymbolUsingNCBI' function the output is showing that GeneID, Symbol and type_of_gene is 'NA'. When I checked the eset data by going into featureDATA and then data, I can see that the data is missing GeneID, Symbol and type_of_gene. How can I fix this problem? Is there something wrong with the way I am loading data into R ? I use raw data from GEO database.
Thanks for help :)
library(limma)
dir()
targets <- readTargets("targets.txt")
targets
#background corection
library(affy)
library(ecolicdf)
#The data is read and normalized using the affy package. The package ecolicdf must also be installed, otherwise the rma() function will attempt to download and install it for you.
eset <- justRMA(filenames = targets$FileName)
colnames(eset) <- row.names(targets)
head(exprs(eset))
plotMDS(eset)
#gene annotation
#First we extract the old gene symbols from the rownames:
Alias <- sub("_.*", "", row.names(eset))
Alias[1:10]
#Then we use limma function alias2SymbolUsingNCBI to update the old gene symbols to current symbols and associated information. The information is stored in the fData slot of the ExpressionSet:
GeneInfo = 'Homo_sapiens.gene_info'
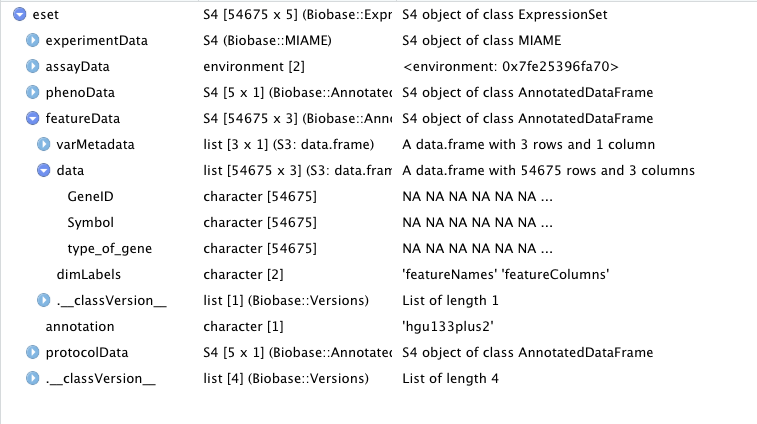
fData(eset) = alias2SymbolUsingNCBI(Alias, GeneInfo, required=c('GeneID', 'Symbol', 'type_of_gene'))
head(fData(eset))
Output of mu code:
library(limma)
> dir()
[1] "GSM1208968_Sample1.genes.fpkm_tracking" "GSM1208968_Sample1.isoforms.fpkm_tracking"
[3] "GSM1208969_Sample2.genes.fpkm_tracking" "GSM1208969_Sample2.isoforms.fpkm_tracking"
[5] "GSM1208970_Sample3.genes.fpkm_tracking" "GSM1208970_Sample3.isoforms.fpkm_tracking"
[7] "GSM1208971_Sample4.genes.fpkm_tracking" "GSM1208971_Sample4.isoforms.fpkm_tracking"
[9] "GSM1208972_Sample5.genes.fpkm_tracking" "GSM1208972_Sample5.isoforms.fpkm_tracking"
[11] "GSM1208973_Sample1.CEL" "GSM1208974_Sample2.CEL"
[13] "GSM1208975_Sample3.CEL" "GSM1208976_Sample4.CEL"
[15] "GSM1208977_Sample5.CEL" "Homo_sapiens.gene_info"
[17] "Project4.Rmd" "Targets.txt"
>
> targets <- readTargets("targets.txt")
> targets
FileName gender source
GSM1208973_Sample1 GSM1208973_Sample1.CEL female second trimester amniotic fluid supernatant
GSM1208974_Sample2 GSM1208974_Sample2.CEL female second trimester amniotic fluid supernatant
GSM1208975_Sample3 GSM1208975_Sample3.CEL male second trimester amniotic fluid supernatant
GSM1208976_Sample4 GSM1208976_Sample4.CEL male second trimester amniotic fluid supernatant
GSM1208977_Sample5 GSM1208977_Sample5.CEL male second trimester amniotic fluid supernatant
>
> #background corection
> library(affy)
> library(ecolicdf)
> #The data is read and normalized using the affy package. The package ecolicdf must also be installed, otherwise the rma() function will attempt to download and install it for you.
> eset <- justRMA(filenames = targets$FileName)
> colnames(eset) <- row.names(targets)
> head(exprs(eset))
GSM1208973_Sample1 GSM1208974_Sample2 GSM1208975_Sample3 GSM1208976_Sample4
1007_s_at 10.229392 10.105533 8.829272 9.581774
1053_at 6.036891 9.086980 6.071632 5.167800
117_at 7.133623 6.398471 6.481419 6.499427
121_at 7.677142 7.092243 7.456217 7.319525
1255_g_at 5.049754 6.201706 8.417677 6.507505
1294_at 6.921347 7.309599 6.921347 7.057446
GSM1208977_Sample5
1007_s_at 8.674735
1053_at 5.781273
117_at 6.472143
121_at 8.314222
1255_g_at 4.924263
1294_at 6.689152
>
> plotMDS(eset)
>
> #gene annotation
> #First we extract the old gene symbols from the rownames:
> Alias <- sub("_.*", "", row.names(eset))
> Alias[1:10]
[1] "1007" "1053" "117" "121" "1255" "1294" "1316" "1320" "1405" "1431"
>
> #Then we use limma function alias2SymbolUsingNCBI to update the old gene symbols to current symbols and associated information. The information is stored in the fData slot of the ExpressionSet:
> GeneInfo = 'Homo_sapiens.gene_info'
> fData(eset) = alias2SymbolUsingNCBI(Alias, GeneInfo, required=c('GeneID', 'Symbol', 'type_of_gene'))
>
> head(fData(eset))
GeneID Symbol type_of_gene
NA <NA> <NA> <NA>
NA.1 <NA> <NA> <NA>
NA.2 <NA> <NA> <NA>
NA.3 <NA> <NA> <NA>
NA.4 <NA> <NA> <NA>
NA.5 <NA> <NA> <NA>


Thank you for your help :D