Hi,
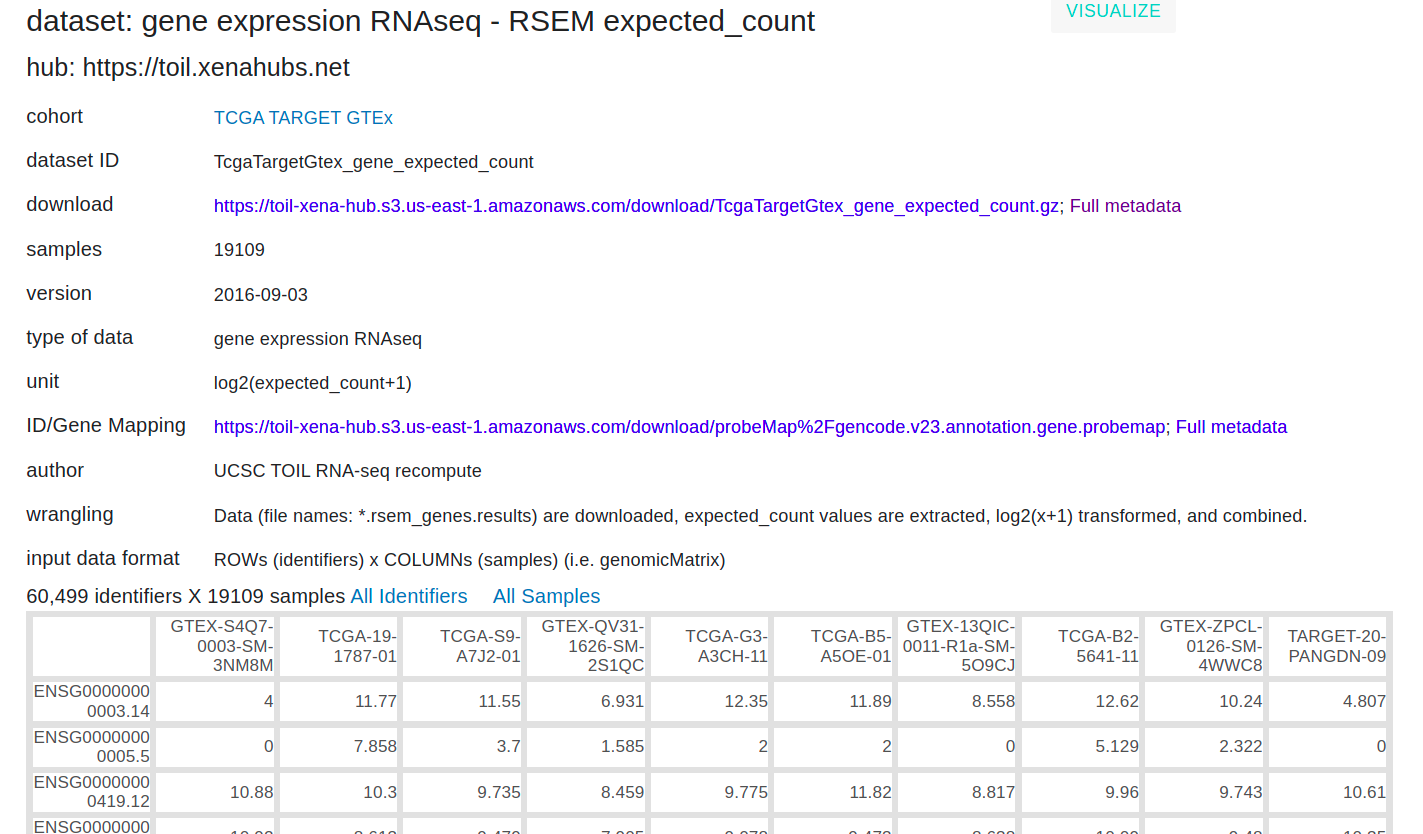
I'm using RNAseq - RSEM expected_count from UCSC RNA-Seq toil recompute data (link to data) for performing differential expression analysis using DESeq2. Details of dataset:
Prior, to performing the de-analysis, the actual counts are computed using 2^(count_from_toil) - 1, and the counts are rounded using the round() function in R.
Please advise if this is the correct approach to be followed.


Michael Love Request your suggestions on this